September in preprints
Posted by the Node, on 3 October 2017
Our latest monthly trawl for developmental biology (and other cool) preprints. See last year’s introductory post for background, and let us know if we missed anything.
This month features butterfly eyespots, brain development in vivo and in silico, lots on cell commitment in embryos and dishes, a diverse selection of modelling preprints, and, right at the end in our ‘Why not…’ section, some algorithmic science art inspired by sand-bubbler crabs!
The preprints were hosted on bioRxiv, PeerJ, arXiv and Wellcome Open Research. Use these links to get to the section you want:
Developmental biology
| Stem cells, regeneration & disease modelling
Evo-devo & evo
Cell biology
Modelling
Tools & resources
| Imaging etc.
Research practice
Why not…
Developmental biology
| Patterning & signalling
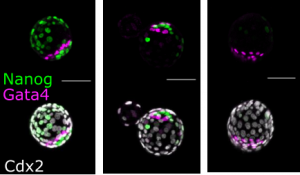
Transcriptional control by Sall4 in blastocysts facilitates lineage commitment of inner cell mass cells. Anzy Miller, Sarah Gharbi, Charles Etienne-Dumeau, Ryuichi Nishinakamura, Brian Hendrich
Dephosphorylation of the NPR2 guanylyl cyclase contributes to inhibition of bone growth by fibroblast growth factor. Leia C. Shuhaibar, Jerid W. Robinson, Ninna P. Shuhaibar, Jeremy R. Egbert, Giulia Vigone, Valentina Baena, Deborah Kaback, Siu-Pok Yee, Robert Feil, Melanie C. Fisher, Caroline N. Dealy, Lincoln R. Potter, Laurinda A. Jaffe
Reconstruction of developmental landscapes by optimal-transport analysis of single-cell gene expression sheds light on cellular reprogramming. Geoffrey Schiebinger, Jian Shu, Marcin Tabaka, Brian Cleary, Vidya Subramanian, Aryeh Solomon, Siyan Liu, Stacie Lin, Peter Berube, Lia Lee, Jenny Chen, Justin Brumbaugh, Philippe Rigollet, Konrad Hochedlinger, Rudolf Jaenisch, Aviv Regev, Eric Lander
MCAM contributes to the establishment of cell autonomous polarity in myogenic and chondrogenic differentiation. Artal Moreno-Fortuny, Laricia Bragg, Giulio Cossu, Urmas Roostalu
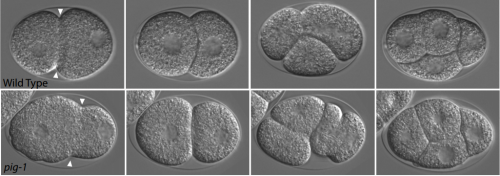
The kinases PIG-1 and PAR-1 act in redundant pathways to regulate asymmetric division in the EMS blastomere of C. elegans. Malgorzata J. Liro, Diane G. Morton, Lesilee S. Rose
In vivo relevance of intercellular calcium signaling in Drosophila wing development. Qinfeng Wu, Pavel Aleksandrovich Brodskiy, Francisco Javier Huizar, Jamison John Jangula, Cody Narciso, Megan Kathleen Levis, Teresa Brito-Robinson, Jeremiah J. Zartman
Intercellular calcium signaling is regulated by morphogens during Drosophila wing development. Pavel A. Brodskiy, Qinfeng Wu, Francisco J. Huizar, Dharsan K. Soundarrajan, Cody Narciso, Megan Levis, Ninfamaria Arredondo-Walsh, Jianxu Chen, Peixian Liang, Danny Z. Chen, Jeremiah James Zartman
The hominoid-specific gene DSCR4 is involved in regulation of human leukocyte migration. Morteza Mahmoudi Saber, Marziyeh Karimiavargani, Nilmini Hettiarachchi, Michiaki Hamada, Takanori Uzawa, Yoshihiro Ito, Naruya Saitou
Ldb1 and Rnf12-dependent regulation of Lhx2 controls the relative balance between neurogenesis and gliogenesis in retina. Jimmy de Melo, Anand Venkataraman, Cristina Zibetti, Brian S. Clark, Seth Blackshaw
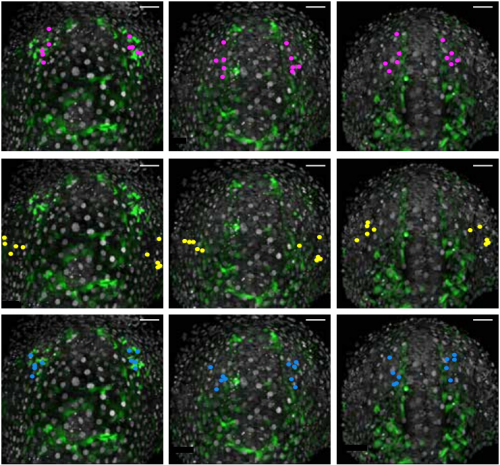
Cell-type heterogeneity in the zebrafish olfactory placode is generated from progenitors within preplacodal ectoderm. Raphaël Aguillon, Julie Batut, Pascale Dufourcq, Romain Madelaine, Arul Subramanian, Thomas F. Schilling, Patrick Blader
Developmental diversification of cortical inhibitory interneurons. Christian Mayer,Christoph Hafemeister, Rachel C Bandler, Robert Machold, Kathryn Allaway, Xavier Jaglin, Renata Batista Brito, Andrew Butler, Gord Fishell, Rahul Satija
Modulation of apoptosis controls inhibitory interneuron number in the cortex. Myrto Denaxa, Guilherme Neves, Adam Rabinowitz, Sarah Kemlo, Petros Liodis, Juan Burrone, Vassilis Pachnis
Paupar LncRNA Promotes KAP1 Dependent Chromatin Changes And Regulates Subventricular Zone Neurogenesis. Ioanna Pavlaki, Farah Alammari, Bin Sun, Neil Clark, Tamara Sirey, Sheena Lee, Dan J. Woodcock, Chris P. Ponting, Francis G. Szele, Keith W. Vance
Single-cell transcriptomic profiling of progenitors of the oligodendrocyte lineage reveals transcriptional convergence during development. Sueli Marques, Darya Vanichkina, David van Bruggen, Elisa Floriddia, Hermany Munguba, Leif Varemo, Stefania Giacomello, Ana Mendanha Falcao, Mandy Meijer, S Samudyata, Simone Codeluppi, Asa Bjorklund, Sten Linnarsson, Jens Hjerling-Leffler, Ryan Taft, Goncalo Castelo-Branco
Embryonic and postnatal neurogenesis produce functionally distinct subclasses of dopaminergic neuron.Elisa Galliano, Eleonora Franzoni, Marine Breton, Annisa N. Chand, Darren J. Byrne, Venkatesh N. Murthy, Matthew S. Grubb
ELMOD1 stimulates ARF6-GTP hydrolysis to stabilize apical structures in developing vestibular hair cells. Jocelyn F Krey, Rachel A Dumont, Philip A Wilmarth, Larry L David, Kenneth R Johnson, Peter G Barr-Gillespie
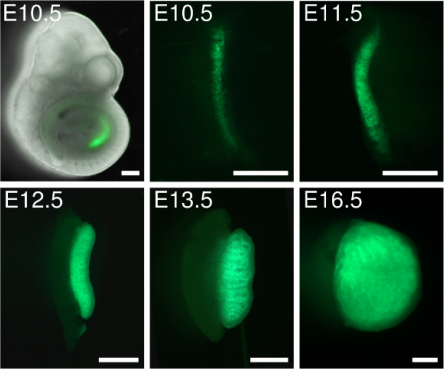
Deciphering cell lineage specification during male sex determination with single-cell RNA sequencing. Isabelle Stevant, Yasmine Neirijnck, Christelle Borel, Jessica Escoffier, Lee B Smith, Stylianos E Antonarakis, Emmanouil T Dermitzakis, Serge Nef
Tissue specific auxin biosynthesis regulates leaf vein patterning. Irina Kneuper, William David Teale, Jonathan Edward Dawson, Ryuji Tsuggeki, Klaus Palme, Eleni Katifori, Franck Anicet Ditengou
Cross-species functional diversity within the PIN auxin efflux protein family. Devin Lee O’Connor, Samuel Elton, Fabrizio Ticchiarelli, Mon Mandy Hsia, John Vogel, Ottoline Leyser
The ERA-related GTPase AtERG2 associated with mitochondria 18S RNA is essential for early embryo development in Arabidopsis. Pengyu Cheng, Hongjuan Li, Linlin Yuan, Huiyong Li, Lele Xi, Junjie Zhang, Jin Liu, Yingdian Wang, Heping Zhao, Huixin Zhao, Shengcheng Han
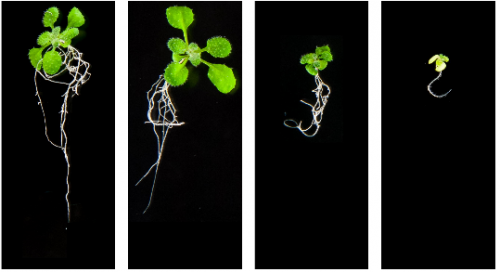
Karrikin-sensing protein KAI2 is a new player in regulating root growth patterns. Stephanie M. Swarbreck, Yannick Guerringue, Elsa Matthus, Fiona J. C. Jamieson, Julia M. Davies
The CLAVATA receptor FASCIATED EAR2 responds to different CLE peptides by signaling through different downstream effectors. Byoung Il Je, Fang Xu, Qingyu Wu, Lei Liu, Robert Meeley, David Jackson
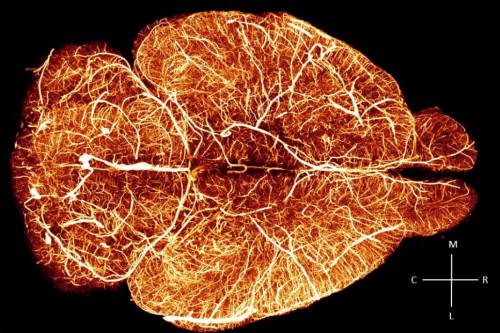
Whole-brain vasculature reconstruction at the single capillary level. Antonino Paolo Di Giovanna, Alessandro Tibo, Ludovico Silvestri, Marie Caroline Muellenbroich, Irene Costantini, Anna Letizia Allegra Mascaro, Leonardo Sacconi, Paolo Frasconi, Francesco Saverio Pavone
Conserved Neural Circuit Structure Across Drosophila Larval Development Revealed By Comparative Connectomics. Stephan Gerhard, Ingrid Andrade, Richard D. Fetter, Albert Cardona, Casey M. Schneider-Mizell
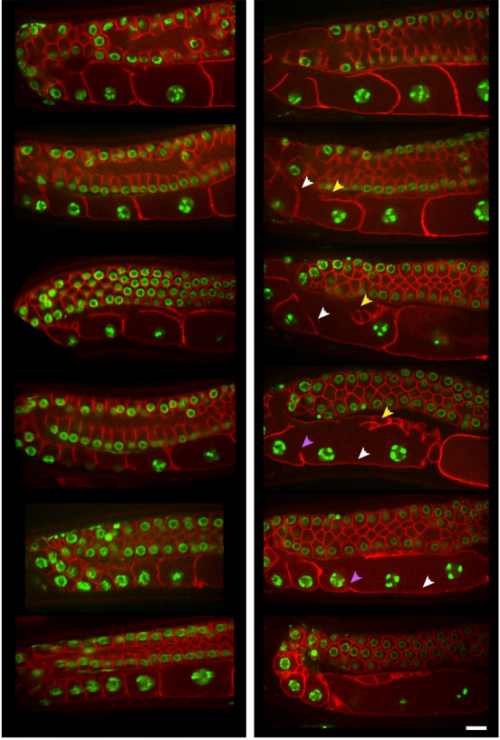
CYK-4 functions independently of its centralspindlin partner ZEN-4 to cellularize oocytes in germline syncytia. Kian-Yong Lee, Rebecca A. Green, Edgar Gutierrez, J. Sebastian Gomez-Cavazos, Irina Kolotuev, Shaohe Wang, Arshad Desai, Alex Groisman, Karen Oegema
Bacterial Colonization Stimulates A Complex Physiological Response In The Immature Human Intestinal Epithelium. David R. Hill, Sha Huang, Melinda S. Nagy, Veda K. Yadagiri, Courtney Fields, Dishari Mukherjee, Brooke Bons, Priya H. Dedhia, Alana M. Chin, Yu-Hwai Tsai, Shrikar Thodla, Thomas M. Schmidt, Seth Walk, Vincent B. Young, Jason R. Spence
C. elegans DBL-1/BMP Regulates Lipid Accumulation via Interaction with Insulin Signaling. James Clark, Michael Meade, Gehan Ranepura, David H Hall, Cathy Savage-Dunn
| Morphogenesis & mechanics
Motility-gradient induced elongation of the vertebrate embryo. Ido Regev, Karine Guevorkian, Olivier Pourquie, L. Mahadevan
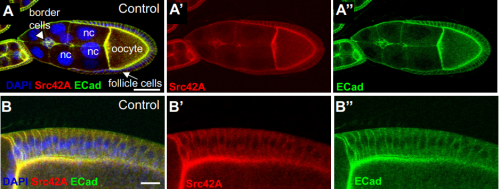
Src42A required for collective border cell migration in vivo. Yasmin Sallak, Alba Yurani Torres, Hongyan Yin, Denise Montell
A “molecular guillotine” reveals an interphase function of Kinesin-5. Zhiyi Lv, Jan Rosenbaum, Timo Aspelmeier, Jorg Grosshans
Drosophila beta-Tubulin 97EF is upregulated at low temperature and stabilizes microtubules. Faina Myachina, Fritz Bosshardt, Johannes Bischof, Moritz Kirschmann, Christian F. Lehner
Microglia remodel synapses by presynaptic trogocytosis and spine head filopodia induction. Laetitia Weinhard, Urte Neniskyte, Giulia di Bartolomei, Giulia Bolasco, Pedro Machado, Nicole Schieber, Melanie Exiga, Auguste Vadisiute, Angelo Raggioli, Andreas Schertel, Yannick Schwab, Cornelius T. Gross
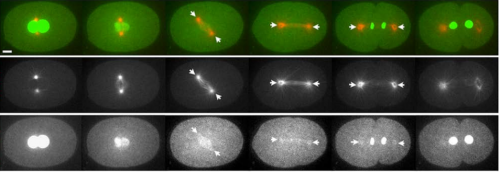
APC/CFZR-1 Controls SAS-5 Levels to Regulate Centrosome Duplication in Caenorhabditis elegans. Jeffrey C. Medley, Lauren E. DeMeyer, Megan M. Kabara, Mi Hye Song
Re-evaluating functional landscape of the cardiovascular system during development. Norio Takada, Madoka Omae, Fumihiko Sagawa, Neil Chi, Satsuki Endo, Satoshi Kozawa, Thomas N Sato
Role of midbody remnant in meiosis II creating tethered polar bodies. Alex McDougall, Celine Hebras, Gerard Pruliere, David Burgess, Vlad Costache, Remi Dumollard, Janet Chenevert
Non-Elastic Remodeling of the 3D Extracellular Matrix by Cell-Generated Forces. Andrea Malandrino, Michael Mak, Xavier Trepat, Roger D. Kamm
Depth Dependent Nanomechanical Analysis of Extracellular Matrix in Multicell Spheroids. Varun Vyas, Melani Solomon, Gerard G. M. D’Souza, Bryan D. Huey
| Genes & genomes
The HoxD Cluster is a Dynamic and Resilient TAD Boundary Controling the Segregation of Antagonistic Regulatory Landscapes. Eddie Rodriguez-Carballo, Lucille Lopez-Delisle, Ye Zhan, Pierre Fabre, Leonardo Beccari, Imane El-Idrissi, Thi Hahn Nguyen Huynh, Hakan Ozadam, Job Dekker, Denis Duboule
Spatially uniform establishment of chromatin accessibility in the early Drosophila embryo. Jenna E. Haines, Michael B. Eisen
dmrad51/spnA mutant exhibit defects during somatic stages of developmental and show enhanced genomic damage, cell death and low temperature sensitivity. Chaitali Khan, Sonia Muliyil, Champakali Ayyub, Basuthkar J. Rao
Chromatin organization changes during the establishment and maintenance of the postmitotic state. Yiqin Ma, Laura Buttitta
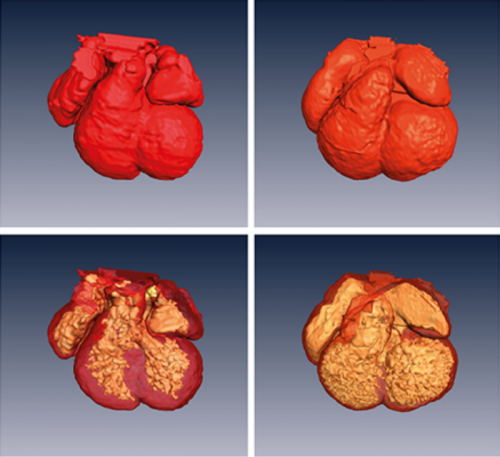
Cardiac enriched BAF chromatin remodeling complex subunit Baf60c regulates gene expression programs essential for heart development and function. Xin Sun, Swetansu Hota, Yu-Qing Zhou, Stefanie Novak, Dario Miguel-Perez, Danos Christodoulou, Christine Seidman, Jonathan Seidman, Carol Gregorio, Mark Henkelman, Janet Rossant, Benoit Bruneau
Nkx2.5-dependent alterations of the embryonic heart DNA methylome identify novel cis-regulatory elements in cardiac development. Bushra Gorsi, Timothy Mosbruger, Megan Smith, Jonathon Hill, H. Joseph Yost
Gene neighbourhood integrity disrupted by CTCF loss in vivo. Dominic Lee, Wilson Tan, George Anene, Peter Li, Tuan Danh, Zenia Tiang, Shi Ling Ng, Motakis Efthymios, Matias Autio, Jianming Jiang, Melissa Fullwood, Shyam Prabhakar, Roger Foo
Nonparametric Bayesian inference of transcriptional branching and recombination identifies regulators of early human germ cell development. Christopher Andrew Penfold, Anastasiya Sybirna, John Reid, Yun Huang, Lorenz Wernisch, Zoubin Ghahramani, Murray Grant, M. Azim Surani
The FACT complex is required for DNA demethylation at heterochromatin during reproduction in Arabidopsis. Jennifer Frost, M. Yvonne Kim, Guen-Tae Park, Ping-Hung Hsieh, Miyuki Nakamura, Samuel Lin, Hyunjin Yoo, Jaemyung Choi, Yoko Ikeda, Tetsu Kinoshita, Yeonhee Choi, Daniel Zilberman, Robert L. Fischer
Genetic Identification of Novel Separase regulators in Caenorhabditis elegans. Michael Melesse, Dillon E. Sloan, Joseph T. Benthal, Quincey Caylor, Krishen Gosine, Xiaofei Bai, Joshua N. Bembenek
Pleiotropy in enhancer function is encoded through diverse genetic architectures. Ella Preger-Ben Noon, Gonzalo Sabarís, Daniela Ortiz, Jonathan Sager, Anna Liebowitz, David L. Stern, Nicolas Frankel
Small RNAs are trafficked from the epididymis to developing mammalian sperm. Upasna Sharma, Fengyun Sun, Brian Reichholf, Veronika Herzog, Stefan Ameres, Oliver Rando
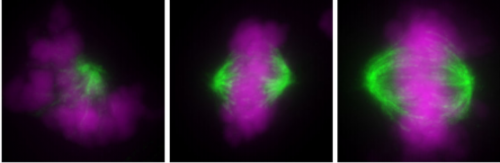
Uncoordinated centrosome duplication cycle underlies the instability of non-diploid states in mammalian somatic cells. Kan Yaguchi, Ryo Matsui, Takahiro Yamamoto, Yuki Tsukada, Atsuko Shibanuma, Keiko Kamimura, Toshiaki Koda, Ryota Uehara
Frequent lack of repressive capacity of promoter DNA methylation identified through genome-wide epigenomic manipulation. Ethan Edward Ford, Matthew R. Grimmer, Sabine Stolzenburg, Ozren Bogdanovic, Alex de Mendoza, Peggy J. Farnham, Pilar Blancafort, Ryan Lister
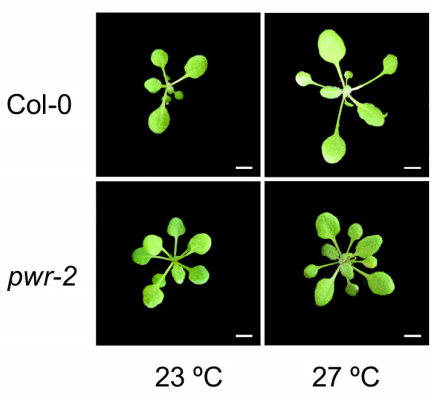
POWERDRESS-mediated histone deacetylation is essential for thermomorphogenesis in Arabidopsis thaliana. Celine Tasset, Avilash Singh Yadav, Sridevi Sureshkumar, Rupali Singh, Lennard van der Woude, Maxim Nekrasov, David Tremethick, Martijn van Zanten, Sureshkumar Balasubramanian
Telomere repeats induce domains of H3K27 methylation in Neurospora. Kirsty Jamieson, Kevin J McNaught, Tereza Ormsby, Neena A. Leggett, Shinji Honda, Eric U Selker
Functional redundancy of variant and canonical histone H3 lysine 9 modification in Drosophila. Taylor J. R. Penke, Daniel J. McKay, Brian D. Strahl, A. Gregory Matera, Robert J. Duronio
NF90/ILF3 is a transcription factor that promotes proliferation over differentiation by hierarchical regulation in K562 erythroleukemia cells. Ting-Hsuan Wu, Lingfang Shi, Jessika Adrian, Minyi Shi, Ramesh V. Nair, Michael P. Snyder, Peter N. Kao
Insights into the molecular changes associated with postnatal human brain development: an integrated transcriptomics and proteomics study. Michael S. Breen, Sureyya Ozcan, Jordan M. Ramsey, Nitin Rustogi, Michael G. Gottshalk, Maree J. Webster, Cyndi Shannon Weickert, Joseph D. Buxbaum, Sabine Bahn
Mating can cause transgenerational gene silencing in Caenorhabditis elegans. Sindhuja Devanapally, Samual Allgood, Antony M. Jose
| Stem cells, regeneration & disease modelling
CATaDa reveals global remodelling of chromatin accessibility during stem cell differentiation in vivo. Gabriel N Aughey, Alicia Estacio Gomez, Jamie Thomson, Hang Yin, Tony D Southall
High-Resolution Dissection of Conducive Reprogramming Trajectory to Ground State Pluripotency. Asaf Zviran, Nofar Mor, Yoach Rais, Hila Gingold, Shani Peles, Elad Chomsky, Sergey Viukov, Jason D. Buenrostro, Leehee Weinberger, Yair S. Manor, Vladislav Krupalnik, Mirie Zerbib, Hadas Hezroni, Diego Adhemar Jaitin, David Larastiaso, Shlomit Gilad, Sima Benjamin, Awni Mousa, Muneef Ayyash, Daoud Sheban, Jonathan Bayerl, Alejandro Aguilera Castrejon, Rada Massarwa, Itay Maza, Suhair Hanna, Ido Amit, Yonatan Stelzer, Igor Ulitsky, William J. Greenleaf, Yitzhak Pilpel, Noa Novershtern, Jacob H. Hanna
SETDB1 prevents TET2-dependent activation of IAP retroelements in naïve embryonic stem cells. Özgen Deniz, Lorenzo de la Rica, Kevin C. L. Cheng, Dominik Spensberger, Miguel R. Branco
Sox4 drives intestinal secretory differentiation toward tuft and enteroendocrine fates. Adam Gracz, Matthew J. Fordham, Danny C. Trotier, Bailey Zwarycz, Yuan-Hung Lo, Katherine Bao, Joshua Starmer, Noah F. Shroyer, Richard L. Reinhardt, Scott T. Magness
FACT sets a barrier for cell fate reprogramming in C. elegans and Human. Ena Kolundzic, Andreas Ofenbauer, Bora Uyar, Anne Sommermeier, Stefanie Seelk, Mei He, Guelkiz Baytek, Altuna Akalin, Sebastian Diecke, Scott Allen Lacadie, Baris Tursun
Polymorphic dynamics of ribosomal proteins gene expression during somatic cell reprogramming and their differentiation into specialized cells-types. Prashanth Kumar Guthikonda, Sumitha Prameela Bharathan, Janakiram Rayabaram, Trinadha Rao Sornapudi, Sailu R Yellaboina, Shaji R Velayudhan, Sreenivasulu Kurukuti
Temporal epigenomic profiling identifies AHR as dynamic super-enhancer controlled regulator of mesenchymal multipotency. Deborah Gérard, Florian Schmidt, Aurélien Ginolhac, Martine Schmitz, Rashi Halder, Peter Ebert, Marcel H. Schulz, Thomas Sauter, Lasse Sinkkonen
Loss of MECP2 leads to induction of p53 and cell senescence. William E Lowry, Minori Ohashi, Peiyee Lee, Kai Fu, Benni Vargas, Denise E. Allen, Elena Korsakova, Jessica K Cinkornpumin, Carlos Salas, Jennifer C Park, Igal Germanguz, Konstantinos Chronis, Edward Kuoy, Stephen Tran, Xinshu Xiao, Matteo Pellegrini, Kathrin Plath
The U2AF homology motif kinase 1 (UHMK1) is upregulated upon hematopoietic cell differentiation. Isabella Barbutti, Joao Agostinho Machado-Neto, Vanessa Cristina Arfelli, Paula de Melo Campos, Fabiola Traina, Sara Teresinha Olalla Saad, Leticia Froehlich Archangelo
Integrated transcriptome and epigenome analyses identify alternative splicing as a novel candidate linking histone modifications to embryonic stem cell fate decision. Yungang Xu, Guangxu Jin, Liang Liu, Dongmin Guo, Xiaobo Zhou
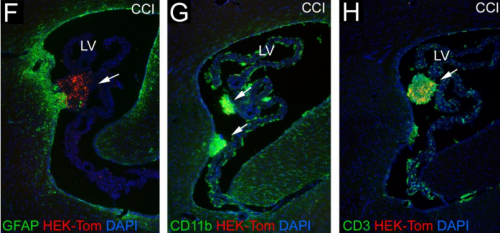
Transplanted Adult Neural Stem Cells Express Sonic Hedgehog In Vivo and Suppress White Matter Neuroinflammation After Experimental Traumatic Brain Injury. Genevieve M. Sullivan, eRegina C. Armstrong
Intraocular injection of ES cell-derived neural progenitors improve visual function in retinal ganglion cell-depleted mouse models. Divya Mundackal Sivaraman, Rasheed Vazhanthodi Abdul, Tiffany Schmidt, Lalitha Soundararajan, Samer Hattar, Jackson James
Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with signatures from post mortem adult brains. Gabriel E. Hoffman, Brigham J. Hartley, Erin Flaherty, Ian Ladran, Peter Gochman, Douglas Ruderfer,Eli A. Stahl, Judith Rapoport, Pamela Sklar, Kristen J. Brennand
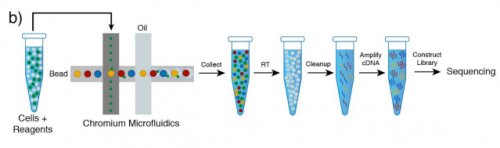
Single Cell RNA Sequencing of stem cell-derived retinal ganglion cells. Maciej Daniszewski, Anne Senabouth, Quan Nguyen, Duncan E Crombie, Samuel W Lukowski, Tejal Kulkarni, Donald J Zack, Alice Pebay, Joseph E Powell, Alex Hewitt
JIP2 haploinsufficiency contributes to neurodevelopmental abnormalities in human pluripotent stem cell-derived neural progenitors and cortical neurons. Reinhard Roessler, Johanna Goldmann, Chikdu Shivalila, Rudolf Jaenisch
LRP1 Regulates Peroxisome Biogenesis and Cholesterol Homeostasis in Oligodendrocytes and is Required in CNS Myelin Development and Repair. Jing-Ping Lin, Yevgeniya A Mironova, Peter Shrager, Roman J Giger
Effects of mechanical loading on cortical defect repair using a novel mechanobiological model of bone healing. Chao Liu, Robert Carrera, Vittoria Flamini, Lena Kenny, Pamela Cabahug-Zuckerman, Benson George, Daniel Hunter, Bo Liu, Gurpreet Singh, Philipp Leucht, Kenneth A Mann, Jill A Helms, Alesha B Castillo
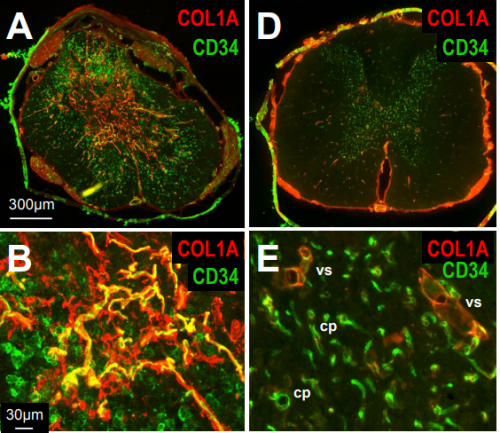
Rats and axolotls share a common molecular signature after spinal cord injury enriched in collagen-1. Athanasios Didangelos, Katalin Bartus, Jure Tica, Michele Puglia, Bernd Roschitzki, Elizabeth Bradbury
DNAJB1-PRKACA fusion kinase drives tumorigenesis and interacts with β-catenin and the liver regenerative response. Edward R Kastenhuber, Gadi Lalazar, Darjus F Tschaharganeh, Shauna L Houlihan, Timour Baslan, Chi-Chao Chen, David Requena, Sha Tian, Benedikt Bosbach, John E Wilkinson, Sanford M Simon, Scott W Lowe
Bcl11b is a Newly Identified Regulator of Vascular Smooth Muscle Phenotype and Arterial Stiffness. Jeff Arni Valisno, Pavania Elavalakanar, Christopher Nicholson, Kuldeep Singh, Dorina Avram, Richard A. Cohen, Gary F. Mitchell, Kathleen G. Morgan, Francesca Seta
Chromatin accessibility profiling uncovers genetic- and T2D disease state-associated changes in cis-regulatory element use in human islets. Shubham Khetan, Romy Kursawe, Ahrim Youn, Nathan Lawlor, Eladio Marquez, Duygu Ucar, Michael L Stitzel
Drosophila larval brain neoplasms present tumour-type dependent genome instability. Fabrizio Rossi, Camille Stephan-Otto Attolini, Jose Luis Mosquera, Cayetano Gonzalez
Pervasive epistasis in cell proliferation pathways modulates neurodevelopmental defects of autism-associated 16p11.2 deletion. Janani Iyer, Mayanglambam Dhruba Singh, Matthew Jensen, Payal Patel, Lucilla Pizzo, Emily Huber, Haley Koerselman, Alexis T. Weiner, Paola Lepanto, Komal Vadodaria, Alexis Kubina, Qingyu Wang, Abigail Talbert, Sneha Yennawar, Jose Badano, J. Robert Manak, Melissa M. Rolls, Arjun Krishnan, Santhosh Girirajan
Targeting histone deacetylase activity to arrest cell growth and promote neural differentiation in Ewing sarcoma. Barbara Kunzler Souza, Patricia Luciana da Costa Lopez, Pamela Rossi Menegotto, Igor Araujo Vieira, Nathalia Kersting, Ana Lucia Abujamra, Andre T. Brunetto, Algemir L. Brunetto, Lauro Gregianin, Caroline Brunetto de Farias, Carol J. Thiele, Rafael Roesler
Fibroblast-derived HGF drives acinar lung cancer cell polarization through integrin-dependent RhoA-ROCK1 inhibition. Anirban Datta, Emma Sandilands, Keith E. Mostov, David M. Bryant
A Rational Drug Combination Design to Inhibit Epithelial-Mesenchymal Transition in a Three-Dimensional Microenvironment. Farnaz Barneh, Mehdi Mirzaie, Payman Nickchi, Tuan Zea Tan, Jean Paul Thiery, Mehran Piran, Mona Salimi, Fatemeh Goshadrou, Amir Reza Aref, Mohieddin Jafari

The age of heterozygous telomerase mutant parents influences the adult phenotype of their offspring irrespective of genotype in zebrafish. Catherine M. Scahill1, Zsofia Digby, Ian M. Sealy, Richard J. White, John E. Collins, Elisabeth M. Busch-Nentwich
Evo-devo & evo
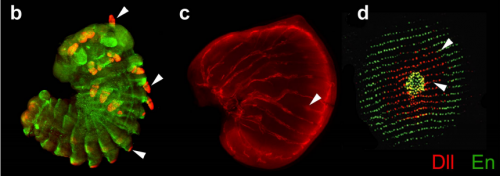
Disrupting different Distal-less exons leads to ectopic and missing eyespots accurately modeled by reaction-diffusion mechanisms. Heidi Connahs, Sham Tlili, Jelle van Creij, Tricia Y. J. Loo, Tirtha Banerjee, Timothy E. Saunders, Antonia Monteiro
The Complex Simplicity of the Brittle Star Nervous System. Olga Zueva, Maleana Khoury, Thomas Heinzeller, Daria Mashanova, Vladimir Mashanov
Early developmental morphology reflects independence from parents in social beetles. Kyle M. Benowitz, Madeline E. Sparks, Elizabeth C. McKinney, Patricia J. Moore, Allen J. Moore
A zombie LIF gene in elephants is up-regulated by TP53 to induce apoptosis in response to DNA damage. Juan Manuel Vazquez, Michael Sulak, Sravanthi Chigurupati, Vincent J. Lynch
Microinjection to deliver protein and mRNA into zygotes of the cnidarian endosymbiosis model Aiptasia sp. Madeline Bucher, Victor Arnold Shivas Jones, Elizabeth Ann Hambleton, Annika Guse
A core signaling mechanism at the origin of animal nociception. Oscar M. Arenas, Emanuela E. Zaharieva, Alessia Para, Constanza Vasquez-Doorman, Christian P. Petersen, Marco Gallio
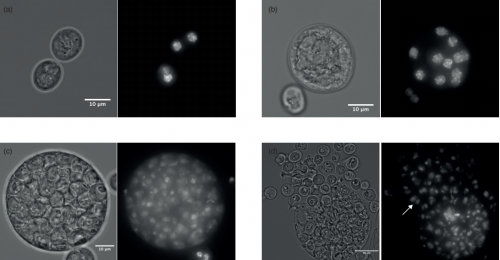
Decoupling of the nuclear division cycle and cell size control in the coenocytic cycle of the ichthyosporean Sphaeroforma arctica. Andrej Ondracka, Iñaki Ruiz-Trillo
Pan-arthropod analysis reveals somatic piRNAs as an ancestral TE defence. Samuel H. Lewis, Kaycee A. Quarles, Yujing Yang, Melanie Tanguy, Lise Frezal, Stephen A. Smith, Prashant P. Sharma, Richard Cordaux, Clement Gilbert, Isabelle Giraud, David H. Collins, Phillip D. Zamore, Eric A. Miska, Peter Sarkies, Francis M. Jiggins
Multispecies coalescent analysis unravels the non-monophyly and controversial relationships of Hexapoda. Lucas A. Freitas, Beatriz Mello, Carlos G. Schrago
Repeat associated mechanisms of genome evolution and function revealed by the Mus caroli and Mus pahari genomes. David Thybert, Maša Roller, Fábio C. P. Navarro, Ian Fiddes, Ian Streeter, Christine Feig, David Martin-Galvez, Mikhail Kolmogorov, Václav Janoušek, Wasiu Akanni, Bronwen Aken, Sarah Aldridge, Varshith Chakrapani, William Chow, Laura Clarke, Carla Cummins, Anthony Doran, Matthew Dunn, Leo Goodstadt, Kerstin Howe, Matthew Howell, Ambre-Aurore Josselin, Robert C. Karn, Christina M. Laukaitis, Lilue Jingtao, Fergal Martin, Matthieu Muffato, Michael A. Quail, Cristina Sisu, Mario Stanke, Klara Stefflova, Cock Van Oosterhout, Frederic Veyrunes, Ben Ward, Fengtang Yang, Golbahar Yazdanifar, Amonida Zadissa, David Adams, Alvis Brazma, Mark Gerstein, Benedict Paten, Son Pham, Thomas Keane, Duncan T. Odom, Paul Flicek
Draft genome of the Eutardigrade Milnesium tardigradum sheds light on ecdysozoan evolution. Felix Mathias Bemm, Laura Burleigh, Frank Foerster, Roland Schmucki, Martin Ebeling, Christian Janzen, Thomas Dandekar, Ralph Schill, Ulrich Certa, Joerg Schultz

Melanism patches up the defective cuticular morphological traits through promoting the up-regulation of cuticular protein-coding genes in Bombyx mori. Liang Qiao, Ri-xin Wang, You-jin Hao, Hai Hu, Gao Xiong, Song-zhen He, Jiang-bo Song, Kun-peng Lu, James Mallet, Ya-qun Xin, Bin Chen, Fang-yin Dai
Genetic architectures of larval pigmentation and color pattern in the redheaded pine sawfly (Neodiprion lecontei). Catherine Linnen, Claire T. O’Quin, Taylor Shackleford, Connor R. Sears, Carita Lindstedt
Conserved microRNA targeting reveals preexisting gene dosage sensitivities that shaped amniote sex chromosome evolution. Sahin Naqvi, Daniel W Bellott, Kathy S Lin, David C Page
Radical changes persist longer in the absence of sex. Joel Sharbrough, Meagan Luse, Jeffrey L Boore, John M Logsdon Jr., Maurine Neiman
Gene transfers, like fossils, can date the Tree of Life. Adrian A. Davin, Eric Tannier, Tom A. Williams, Bastien Boussau, Vincent Daubin, Gergely J. Szollosi
Virtual Genome Walking: Generating gene models for the salamander Ambystoma mexicanum. Teri Evans, Andrew D. Johnson, Matthew D. Loose
Conflict between heterozygote advantage and hybrid incompatibility in haplodiploids (and sex chromosomes). Ana-Hermina Ghenu, Alexandre Blanckaert, Roger K. Butlin, Jonna Kulmuni, Claudia Bank
EVOLUTIONARY ANALYSIS OF CANDIDATE NON-CODING ELEMENTS REGULATING NEURODEVELOPMENTAL GENES IN VERTEBRATES. Francisco J. Novo
Dramatic evolution of body length due to post-embryonic changes in cell size in a newly discovered close relative of C. elegans. Gavin C. Woodruff, Patrick C. Phillips
The crowns have eyes: Multiple opsins found in the eyes of the Crown-of-Thorns Starfish Acanthaster planci including the first r-opsin utilized by a deuterostome eye. Elijah K. Lowe, Anders Garm, Esther Ullrich-Luter, Maria Ina Arnone
Evidence for “inter- and intraspecific horizontal genetic transfers” between anciently asexual bdelloid rotifers is explained by cross-contamination. Christopher G Wilson, Reuben W Nowell, Timothy G Barraclough
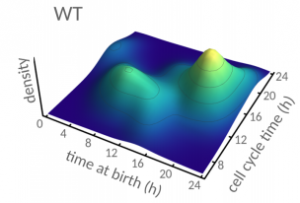
Cell size control driven by the circadian clock and environment in cyanobacteria. Bruno M. C. Martins, Amy K. Tooke, Philipp Thomas, James C. W. Locke
Cell biology
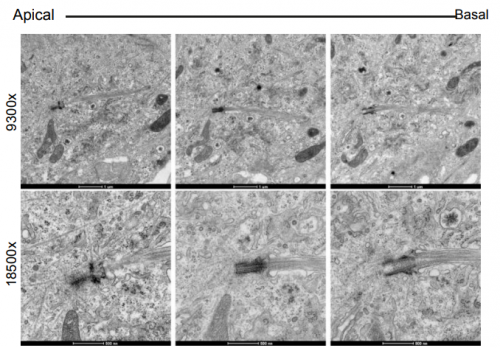
Regulator of calcineurin-2 is a ciliary protein with a role in cilia length control. Nicola Stevenson, Dylan Bergen, Amadeus Xu, Emily Wyatt, Freya Henry, Janine McCaughey, Laura Vuolo, Chrissy Hammond, David Stephens
Dynamics of the IFT Machinery at the Ciliary Tip. Alexander Chien, Sheng Min Shih, Raqual Bower, Douglas Tritschler, Mary E. Porter, Ahmet Yildiz
Dynamic Spatiotemporal Organization of Exocytosis During Cellular Shape Change. Fabio Urbina, Shawn Gomez, Stephanie L. Gupton
Microtubule regulation of integrin-based adhesions is mediated by myosin-IIA. Yukako Nishimura, Nisha Mohd Rafiq, Sergey V. Plotnikov, Zhen Zhang, Visalatchi Thiagarajan, Meenubharathi Natarajan, Gareth E. Jones, Pakorn Kanchanawong, Alexander D. Bershadsky
Force dependence of filopodia adhesion: involvement of myosin II and formins. Naila O. Alieva, Artem K. Efremov, Shiqiong Hu, Dongmyung Oh, Zhongwen Chen, Meenubharathi Natarajan, Hui Ting Ong, Antoine Jegou, Guillaume Romet-Lemonne, Jay T. Groves, Michael P. Sheetz, Jie Yan, Alexander D. Bershadsky
The lamellipodium is a myosin independent mechanosensor. Patrick W. Oakes, Tamara C. Bidone, Yvonne Beckham, Austin V. Skeeters, Guillermina R. Ramirez-San Juan, Stephen P. Winter, Gregory A. Voth, Margaret L. Gardel
Architecture of mammalian centriole distal appendages accommodates distinct blade and matrix functional elements. T. Tony Yang, Weng Man Chong, Won-Jing Wang, Gregory Mazo, Barbara Tanos, Zhengmin Chen, Minh Nguyet Thi Tran, Yi-De Chen, Rueyhung Roc Weng, Chia-En Huang, Wann-Neng Jane, Meng-Fu Bryan Tsou, Jung-Chi Liao
Spatial and temporal translocation of PKCα in single endothelial cell in response to focal mechanical stimulus. Masataka Arai, Toshihiro Sera, Takumi Hasegawa, Susumu Kudo
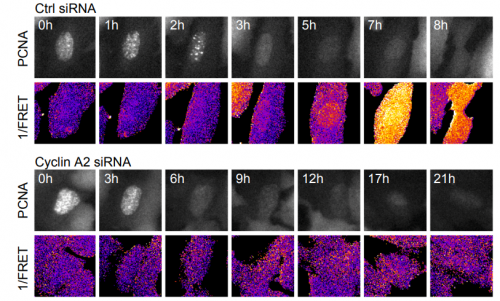
Cyclin A2 localises in the cytoplasm at the S/G2 transition to activate Plk1. Helena Silva Cascales, Kamila Burdova, Erik Mullers, Henriette Stoy, Patrick von Morgen, Libor Macurek, Arne Lindqvist
CDK1 and PLK1 co-ordinate the disassembly and re-assembly of the Nuclear Envelope in vertebrate mitosis. Ines J de Castro, Raquel Sales Gil, Lorena Ligammari, Maria Laura Di Giacinto, Paola Vagnarelli
Phosphatase PP2A and microtubule pulling forces disassemble centrosomes during mitotic exit. Stephen J. Enos, Martin Dressler, Beatriz Ferreira Gomes, Anthony A. Hyman, Jeffrey B. Woodruff
Resolving ESCRT-III spirals at the intercellular bridge of dividing cells using 3D STORM imaging. Inna Goliand, Tali Dadosh, Natalie Elia
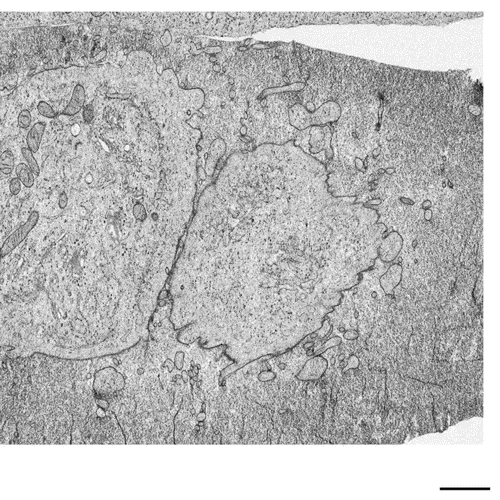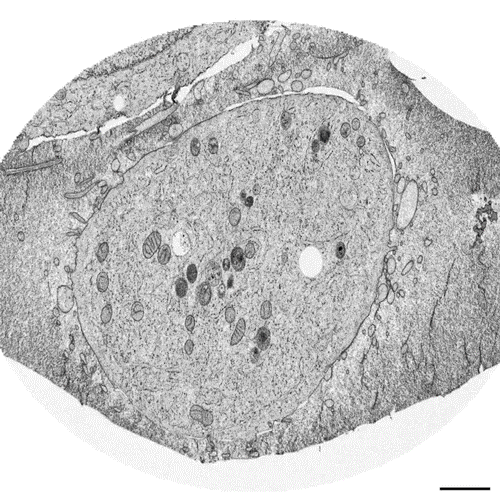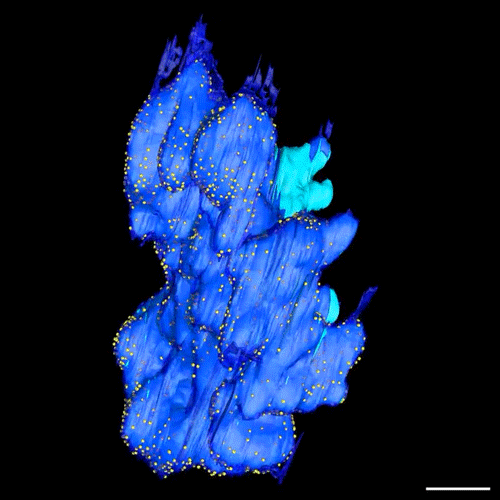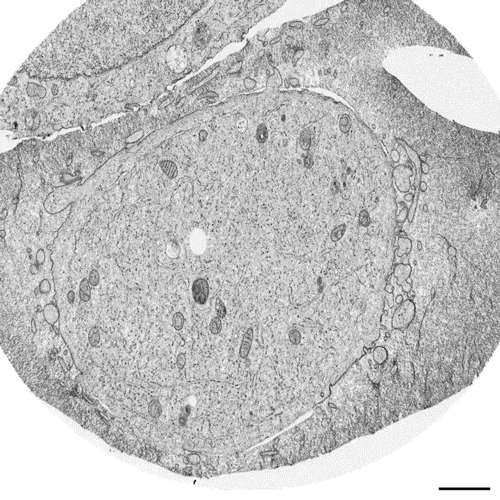
Postmitotic Nuclear Pore Assembly Proceeds By Radial Dilation Of Small ER Membrane Openings. Shotaro Otsuka, Anna M. Steyer, Martin Schorb, Jean-Karim Heriche, M. Julius Hossain, Suruchi Sethi, Moritz Kueblbeck, Yannick Schwab, Martin Beck, Jan Ellenberg
Nano-scale size holes in ER sheets provide an alternative to tubules for highly-curved membranes. Lena K. Schroeder, Andrew E.S. Barentine, Sarah Schweighofer, David Baddeley, Joerg Bewersdorf, Shirin Bahmanyar
Human replication licensing factor Cdt1 directly links mitotic kinetochores to spindle microtubules. Shivangi Agarwal, Kyle Smith, Yizhuo Zhou, Aussie Suzuki, Richard McKenney, Dileep Varma
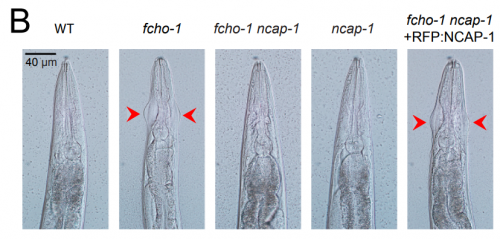
NECAPs are negative regulators of the AP2 clathrin adaptor complex. Gwendolyn M Beacham, Edward A Partlow, Jeffrey J Lange, Gunther Hollopeter
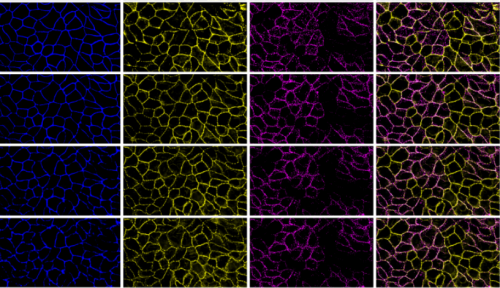
Propagating actomyosin-generated force to intercellular junction. Vivian W. Tang
Distinct prophase arrest mechanisms in human male meiosis. Sabrina Z. Jan, Aldo Jongejan, Cindy M. Korver, Saskia K. M. van Daalen, Ans M. M. van Pelt, Sjoerd Repping, Geert Hamer
ATR is a multifunctional regulator of male mouse meiosis. Alexander Widger, Shantha K Mahadevaiah, Julian Lange, Elias ElInati, Jasmin Zohren, Takayuki Hirota, Marcello Stanzione, Obah Ojarikre, Valdone Maciulyte, Dirk de Rooij, Attila Toth, Scott Keeney, James MA Turner
Non-canonical circadian oscillations in Drosophila S2 cells drive gene-expression cycles coupled to metabolic oscillations. Guillaume Rey, Nikolay B. Milev, Utham K. Valekunja, Ratnasekhar Ch, Sandipan Ray, Mariana Silva Dos Santos, Andras D. Nagy, Robin Antrobus, James I. MacRae, Akhilesh B. Reddy
The Cell-Cycle Transcriptional Network Generates and Transmits a Pulse of Transcription Once Each Cell Cycle. Chun-Yi Cho, Christina M. Kelliher, Steven B. Haase
Cell contraction induces long-ranged stress stiffening in the extracellular matrix. Yu Long Han, Pierre Ronceray, Guoqiang Xu, Andrea Malandrino, Roger Kamm, Martin Lenz, Chase P. Broedersz, Ming Guo
Modelling
Approximate Bayesian computation reveals the importance of repeated measurements for parameterising cell-based models of growing tissues. Jochen Kursawe, Ruth E. Baker, Alexander George Fletcher
Inferring parameters for a lattice-free model of cell migration and proliferation using experimental data. Alexander Browning, Scott W. McCue, Rachelle N. Binny, Michael J. Plank, Esha T. Shah, Matthew J. Simpson
Reprogramming, oscillations and transdifferentiation in epigenetic landscapes. Bivash Kaity, Ratan Sarkar, Buddhapriya Chakrabarti, Mithun K. Mitra
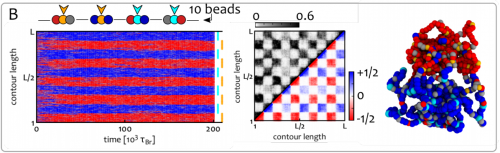
Shaping Epigenetic Memory via Genomic Bookmarking. Davide Michieletto, Michael Chiang, Davide Coli, Argyris Papantonis, Enzo Orlandini, Peter R. Cook, Davide Marenduzzo
Optimal quantification of contact inhibition in cell populations. David J. Warne, Ruth E. Baker, Matthew J. Simpson
In silico mechanobiochemical modeling of morphogenesis in cell monolayers. Hongyan Yuan, Bahador Marzban
Bifurcations in valveless pumping techniques from a coupled fluid-structure-electrophysiology model in heart development. Nicholas A. Battista, Laura A. Miller
Theory of epithelial cell shape transitions induced by mechanoactive chemical gradients. Kinjal Dasbiswas, Edouard Hannezo, Nir. S. Gov
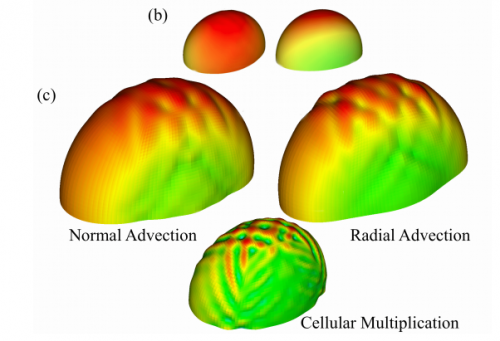
A computational study of growth-driven folding patterns on shells, with application to the developing brain. S. N. Verner, K. Garikipati
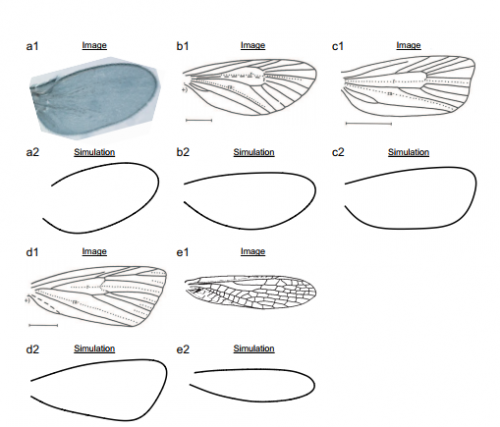
A mechanical model for diversified insect wing margin shapes. Yukitaka Ishimoto, Kaoru Sugimura
Analysis of biochemical mechanisms provoking differential spatial expression in Hh target genes. Manuel Cambón
The importance of geometry in the corneal micropocket assay. James A Grogan, Anthony J Connor, Joe M Pitt-Francis, Philip K Maini, Helen M Byrne
Functional Modeling of Plant Growth Dynamics. Yuhang Xu, Yumou Qiu, James Schnable
Integration of anatomy ontologies and Evo-Devo using structured Markov chains suggests a new framework for modeling discrete phenotypic traits. Sergei Tarasov
Spatial cytoskeleton organization supports targeted intracellular transport. Anne E. Hafner, Heiko Rieger
Consistent Reanalysis of Genome-wide Imprinting Studies in Plants Using Generalized Linear Models Increases Concordance across Datasets. Stefan Wyder, Michael T Raissig, Ueli Grossniklaus
Replication Timing Networks: a novel class of gene regulatory networks. Juan Carlos Rivera-Mulia, Sebo Kim, Haitham Gabr, Tamer Kahveci, David M Gilbert
Tools & resources
| Imaging etc.
Low cost and open source multi-fluorescence imaging system for teaching and research in biology and bioengineering. Isaac Nuñez, Tamara Matute, Roberto Herrera, Juan Keymer, Tim Marzullo, Tim Rudge, Fernan Federici
PhysiCell: an Open Source Physics-Based Cell Simulator for 3-D Multicellular Systems. Ahmadreza Ghaffarizadeh, Samuel H. Friedman, Shannon M. Mumenthaler, Paul Macklin
Hemodynamic forces can be accurately measured in vivo with optical tweezers. Sebastien Harlepp, Fabrice Thalmann, Gautier Follain, Jacky G. Goetz
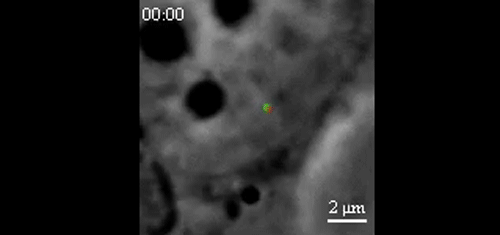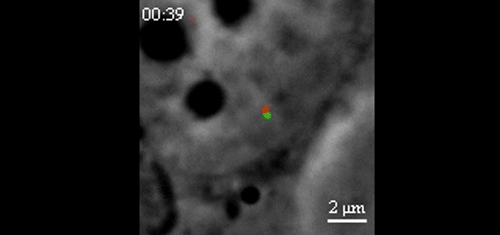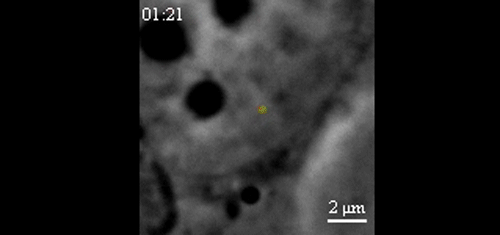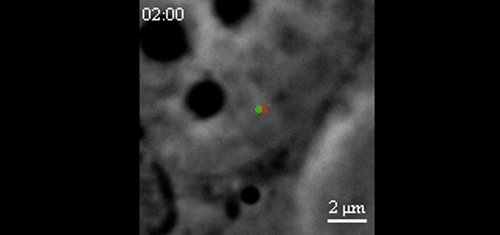
CRISPR-Based DNA Imaging in Living Cells Reveals Cell Cycle-Dependent Chromosome Dynamics. Hanhui Ma, Li-Chun Tu, Ardalan Naseri, Yu-Chieh Chung, David Grunwald, Shaojie Zhang, Thoru Pederson
Simultaneously measuring image features and resolution in live-cell STED images. Andrew E. S. Barentine, Lena K. Schroeder, Michael Graff, David Baddeley, Joerg Bewersdorf
Obtaining 3D Super-resolution Information from 2D Super-resolution Images through a 2D-to-3D Transformation Algorithm. Andrew Ruba, Joseph Kelich, Wangxi Luo, Weidong Yang
Hydrogel encapsulation of living organisms for long-term microscopy. Kyra Burnett, Eric Edsinger, Dirk R. Albrecht
Quantification of cellular distribution as Poisson process in 3D matrix using a multiview light-sheet microscope. Warren Colomb, Matthew Osmond, Charles Durfee, Melissa D. Krebs, Susanta K. Sarkar
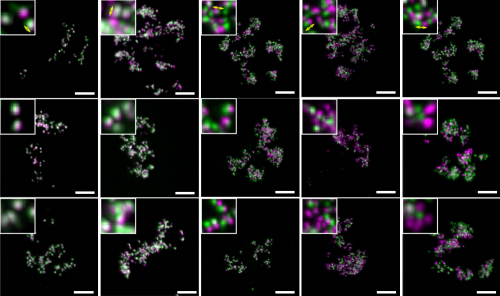
Correlative live and super-resolution imaging reveals the dynamic structure of replication domains. Wanqing Xiang, M. Julia Roberti, Jean-Karim Heriche, Sebastian Huet, Stephanie Alexander, Jan Ellenberg
Coupling optogenetics and light-sheet microscopy to study signal transduction in vivo. Prameet Kaur, Timothy E. Saunders, Nicholas Stanislaw Tolwinski
Arabidopsis phenotyping through Geometric Morphometrics. Carlos A. Manacorda, Sebastian Asurmendi
MICCS: A Fully Programmable Multipurpose Integrated Cell Culture System. Timothy Kassis, Paola M. Perez, Chloe J. W. Yang, Luis R. Soenksen, David L. Trumper, Linda G. Griffith
vU-net: accurate cell edge segmentation in time-lapse fluorescence live cell images based on convolutional neural network. Chuangqi Wang, Xitong Zhang, Yenyu Chen, Kwonmoo Lee
ChromoTrace: Reconstruction of 3D Chromosome Configurations by Super-Resolution Microscopy. Carl Barton, Sandro Morganella, Oeyvind Oedegaard, Stephanie Alexander, Jonas Ries, Tomas Fitzgerald, Jan Ellenberg, Ewan Birney
PlantCV v2.0: Image analysis software for high-throughput plant phenotyping. Malia A Gehan, Noah Fahlgren, Arash Abbasi, Jeffrey C Berry, Steven T Callen, Leonardo Chavez, Andrew N Doust, Max J Feldman, Kerrigan B Gilbert, John G Hodge, J Steen Hoyer, Andy Lin, Suxing Liu, César Lizárraga, Argelia Lorence, Michael Miller, Eric Platon, Monica Tessman, Tony Sax

Raspberry Pi Powered Imaging for Plant Phenotyping. Jose Tovar, John Steen Hoyer, Andy Lin, Allison Tielking, Steven Callen, Elizabeth Castillo, Michael Miller, Monica Tessman, Noah Fahlgren, James Carrington, Dmitri Nusinow, Malia A. Gehan
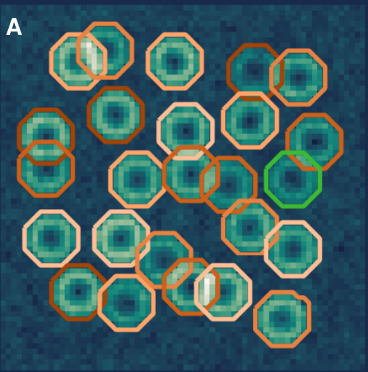
ABLE: an Activity-Based Level Set Segmentation Algorithm for Two-Photon Calcium Imaging Data. Stephanie Reynolds, Therese Abrahamsson, Renaud Schuck, P. Jesper Sjöström, Simon R Schultz, Pier Luigi Dragotti
Edge Detection of Cryptic Lamellipodia Assisted by Deep Learning. Chuangqi Wang, Shawn Kang, Eunice Kim, Xitong Zhang, Hee June Choi, Aaron Choi, Kwonmoo Lee
Mapping nonapoptotic caspase activity with a transgenic reporter in mice. Peter Nicholls, Thomas Pack, Nikhil Urs, Sunil Kumar, Gabor Turu, Evan Calabrese, Wendy Roberts, Ping Fan, Valeriy Ostapchenko, Monica Guzman, Flavio Beraldo, Vania Prado, Marco Prado, Ivan Spasojevic, Joshua Snyder, Kafui Dzirasa, G. Allan Johnson, Marc Caron
Development of an Intrinsic Skin Sensor for Blood Glucose Level with CRISPR-mediated Genome Editing in Epidermal Stem Cells. Jiping Yue, Yuanyuan Li, Xuewen Gou, Xiaoyang Wu
Mini-ring approach for high-throughput drug screenings in 3D tumor models. Nhan Phan, Bobby Tofig, Deanna Janzen, Jin Huang, Sanaz Memarzadeh, Robert Damoiseaux, Alice Soragni
A Cautionary Tail: Changes in Integrin Behavior with Labeling. Catherine G. Galbraith, Michael W. Davidson, James A. Galbraith
| Genome tools
Employing single-stranded DNA donors for the high-throughput production of conditional knockout alleles in mice. Denise G. Lanza, Angelina Gaspero, Isabel Lorenzo, Lan Liao, Ping Zheng, Ying Wang, Yu Deng, Chonghui Cheng, Chuansheng Zhang, Matthew N. Rasband, John R. Seavitt, Francisco J. DeMayo, Jianming Xu, Mary E. Dickinson, Arthur L. Beaudet, Jason D. Heaney
Generation and validation of homozygous fluorescent knock-in cells using genome editing. Birgit Koch, Bianca Nijmeijer, Moritz Kueblbeck, Yin Cai, Nike Walther, Jan Ellenberg
Precision genome-editing with CRISPR/Cas9 in human induced pluripotent stem cells. John P. Budde, Rita Martinez, Simon Hsu, Natalie Wen, Jason A. Chen, Giovanni Coppola, Alison M. Goate, Carlos Cruchaga, Celeste Karch
Unexpected CRISPR off-target mutation pattern in vivo are not typically germline-like. Zhiting Wei, Funan He, Guohui Chuai, Hanhui Ma, Zhixi Su, Qi Liu
Crossing enhanced and high fidelity SpCas9 nucleases to optimize specificity and cleavage. Peter Istvan Kulcsar, Andras Talas, Krisztina Huszar, Zoltan Ligeti, Eszter Toth, Nora Weinhardt, Elfrieda Fodor, Ervin Welker
High-throughput creation and functional profiling of eukaryotic DNA sequence variant libraries using CRISPR/Cas9. Xiaoge Guo, Alejandro Chavez, Angela Tung, Yingleong Chan, Ryan Cecchi, Santiago Lopez Garnier, Christian Kaas, Eric Kelsic, Max Schubert, James DiCarlo, James Collins, George Church
FlashFry: a fast and flexible tool for large-scale CRISPR target design. Aaron McKenna, Jay Shendure
indCAPS: A tool for designing screening primers for CRISPR/Cas9 mutagenesis events. Charles Hodgens, Zachary Nimchuk, Joseph Kieber
Integrated design, execution, and analysis of arrayed and pooled CRISPR genome editing experiments. Matthew C. Canver, Maximilian Haeussler, Daniel E. Bauer, Stuart H. Orkin, Neville E. Sanjana, Ophir Shalem, Guo-Cheng Yuan, Feng Zhang, Jean-Paul Concordet, Luca Pinello
Mechanisms of improved specificity of engineered Cas9s revealed by single molecule analysis. Digvijay Singh, Yanbo Wang, John Mallon, Olivia Yang, Jingyi Fei, Anustup Poddar, Damon Ceylan, Scott Bailey, Taekjip Ha
Characterization and Validation of a Novel Group of Type V, Class 2 Nucleases for in vivo Genome Editing. Matthew B Begemann, Benjamin N Gray, Emma January, Anna Singer, Dylan C Kesler, Yonghua He, Haijun Liu, Hongjie Guo, Alex Jordan, Thomas P Brutnell, Todd C Mockler, Mohammed Oufattole
High-resolution genome-wide functional dissection of transcriptional regulatory regions in human. Xinchen Wang, Liang He, Sarah Goggin, Alham Saadat, Li Wang, Melina Claussnitzer, Manolis Kellis
Multi-platform discovery of haplotype-resolved structural variation in human genomes. Mark J.P. Chaisson, Ashley D. Sanders, Xuefang Zhao, Ankit Malhotra, David Porubsky, Tobias Rausch, Eugene J. Gardner, Oscar Rodriguez, Li Guo, Ryan L. Collins, Xian Fan, Jia Wen, Robert E. Handsaker, Susan Fairley, Zev N. Kronenberg, Xiangmeng Kong, Fereydoun Hormozdiari, Dillon Lee, Aaron M. Wenger, Alex Hastie, Danny Antaki, Peter Audano, Harrison Brand, Stuart Cantsilieris, Han Cao, Eliza Cerveira, Chong Chen, Xintong Chen, Chen-Shan Chin, Zechen Chong, Nelson T. Chuang, Deanna M. Church, Laura Clarke, Andrew Farrell, Joey Flores, Timur Galeev, Gorkin David, Madhusudan Gujral, Victor Guryev, William Haynes-Heaton, Jonas Korlach, Sushant Kumar, Jee Young Kwon, Jong Eun Lee, Joyce Lee, Wan-Ping Lee, Sau Peng Lee, Patrick Marks, Karine Valud-Martinez, Sascha Meiers, Katherine M. Munson, Fabio Navarro, Bradley J. Nelson, Conor Nodzak, Amina Noor, Sofia Kyriazopoulou-Panagiotopoulou, Andy Pang, Yunjiang Qiu, Gabriel Rosanio, Mallory Ryan, Adrian Stutz, Diana C.J. Spierings, Alistair Ward, AnneMarie E. Welsch, Ming Xiao, Wei Xu, Chengsheng Zhang, Qihui Zhu, Xiangqun Zheng-Bradley, Goo Jun, Li Ding, Chong Lek Koh, Bing Ren, Paul Flicek, Ken Chen, Mark B. Gerstein, Pui-Yan Kwok, Peter M. Lansdorp, Gabor Marth, Jonathan Sebat, Xinghua Shi, Ali Bashir, Kai Ye, Scott E. Devine, Michael Talkowski, Ryan E. Mills, Tobias Marschall, Jan Korbel, Evan E. Eichler, Charles Lee
Successful optimization of CRISPR/Cas9-mediated defined point mutation knock-in using allele-specific PCR assays in zebrafish. Sergey V. Prykhozhij, Charlotte Fuller, Shelby L. Steele, Chansey J. Veinotte, Babak Razaghi, Christopher McMaster, Adam Shlien, David Malkin, Jason N. Berman
Profiling DNA Methylation Differences Between Inbred Mouse Strains on the Illumina Human Infinium MethylationEPIC Microarray. Hemant Gujar, Jane W Liang, Nicholas C Wong, Khyobeni Mozhui
From trash to treasure: detecting unexpected contamination in unmapped NGS data. llaria Granata, Mara Sangiovanni, Amarinder Singh Thind, Mario Rosario Guarracino.
Accurate estimation of molecular counts in droplet-based single-cell RNA-seq experiments. Viktor Petukhov, Jimin Guo, Ninib Baryawno, Nicolas Severe, David Scadden, Maria G. Samsonova, Peter V. Kharchenko
Gene synthesis allows biologists to source genes from farther away in the tree of life. Aditya Kunjapur, Philipp Pfingstag, Neil C Thompson
Curated compendium of human transcriptional biomarker data. Nathan P Golightly, Anna I Bischoff, Avery Bell, Parker D Hollingsworth, Stephen R Piccolo
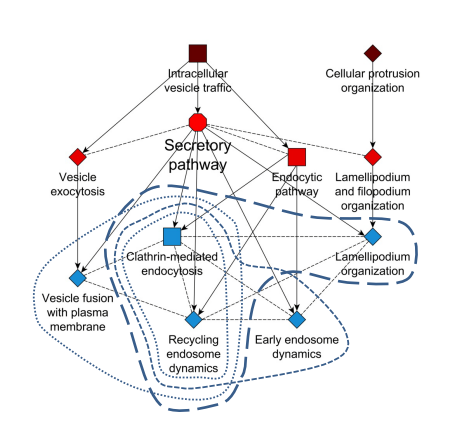
A flexible ontology for inference of emergent whole cell function from relationships between subcellular processes. Jens Hansen, David Meretzky, Simeneh Woldesenbet, Gustavo Stolovitzky, Ravi Iyengar
Comparison of seven single cell Whole Genome Amplification commercial kits using targeted sequencing. Tamir Biezuner, Ofir Raz, Shiran Amir, Lilach Milo, Rivka Adar, Yael Fried, Elena Ainbinder, Ehud Shapiro
Biosensor libraries harness large classes of binding domains for allosteric transcription regulators. Javier F. Juarez, Begoña Lecube-Azpeitia, Stuart L. Brown, George M. Church
An automated model test system for systematic development and improvement of gene expression models. Alexander C Reis, Howard Salis
Using DNase Hi-C techniques to map global and local three-dimensional genome architecture at high resolution. Wenxiu Ma, Ferhat Ay, Choli Lee, Gunhan Gulsoy, Xinxian Deng, Savannah Cook, Jennifer Hesson, Christopher Cavanaugh, Carol B. Ware, Anton Krumm, Jay Shendure, C. Anthony Blau, Christine M. Disteche, William S. Noble, Zhijun Duan
Massive Mining of Publicly Available RNA-seq Data from Human and Mouse. Alexander Lachmann, Denis Torre, Alexandra B. Keenan, Kathleen M. Jagodnik, Hyojin J. Lee, Moshe C. Silverstein, Lily Wang, Avi Ma’ayan
LeafCutter: Annotation-free quantification of RNA splicing. Yang I. Li, David A. Knowles, Jack Humphrey, Alvaro N. Barbeira, Scott P. Dickinson, Hae Kyung Im, Jonathan K. Pritchard
A sequence-based, deep learning model accurately predicts RNA splicing branchpoints. Joseph M. Paggi, Gill Bejerano
Superior ab initio Identification, Annotation and Characterisation of TEs and Segmental Duplications from Genome Assemblies. LU ZENG, R Daniel Kortschak, Joy M Raison, Terry Bertozzi, David L Adelson
DeepBlueR: Large-scale epigenomic analysis in R. Markus List, Felipe Albrecht, Christoph Bock, Thomas Lengauer
mosdepth: quick coverage calculation for genomes and exomes. Brent S. Pedersen, Aaron Quinlan
CUT&RUN: Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Peter J. Skene, Steven Henikoff
Real-time fluorescence and deformability cytometry – flow cytometry goes mechanics. Philipp Rosendahl, Katarzyna Plak, Angela Jacobi, Martin Kraeter, Nicole Toepfner, Oliver Otto, Christoph Herold, Maria Winzi, Maik Herbig, Yan Ge, Salvatore Girardo, Katrin Wagner, Buzz Baum, Jochen Guck
DAFi: A Directed Recursive Filtering and Clustering Approach to Data-Driven Identification of Cell Populations from Polychromatic Flow Cytometry Data. Alexandra J Lee, Ivan Chang, Julie G Burel, Cecilia S Lindestam Arlehamn, Daniela Weiskopf, Bjoern Peters, Alessandro Sette, Richard H Scheuermann, Yu Qian
Interactive Visual Analysis of Mass Cytometry Data by Hierarchical Stochastic Neighbor Embedding Reveals Rare Cell Types. Vincent van Unen, Thomas Hollt, Nicola Pezzotti, Na Li, Marcel J. T. Reinders, Elmar Eisemann, Anna Vilanova, Frits Koning, Boudewijn P. F. Lelieveldt
HyperTRIBE: Upgrading TRIBE with enhanced editing. Weijin Xu, Reazur Rahman, Michael Rosbash
plot2DO: a tool to assess the quality and distribution of genomic data. Răzvan V Chereji
Gene regulatory network inference from single-cell data using multivariate information measures. Thalia E Chan, Michael Stumpf, Ann C Babtie
A duplex MIPs-based biological-computational cell lineage discovery platform. Liming Tao, Ofir Raz, Zipora Marx, Tamir Biezuner, Shiran Amir, Lilach Milo, Rivka Adar, Amos Onn, Noa Chapal-Ilani, Veronika Berman, Ron Levy, Barak Oron, Ehud Shapiro
Deep learning of the splicing (epi)genetic code reveals a novel candidate mechanism linking histone modifications to ESC fate decision. Yungang Xu, Yongcui Wang, Jiesi Luo, Weiling Zhao, Xiaobo Zhou
miRNAgFree: prediction and profiling of novel microRNAs without genome assembly. Ernesto L. Aparicio, Antonio Rueda, Bastian Fromm, Cristina Gómez-Martin, Ricardo Lebrón, Jose L Oliver, Juan A Marchal, Michail Kotsyfakis, Michael Hackenberg
Trendy: Segmented regression analysis of expression dynamics for high-throughput ordered profiling experiments. Rhonda Bacher, Ning Leng, Li-Fang Chu, James Thomson, Christina Kendziorski, Ron Stewart
MTopGO: a tool for module identification in PPI Networks. Danila Vella, Simone Marini, Francesca Vitali, Riccardo Bellazzi
Research practice
Award or Reward? Which comes first, NIH funding or research impact? Lucas C Parra, Lukas Hirsch
Assessment of the impact of shared data on the scientific literature. Michael Milham, Cameron Craddock, Michael Fleischmann, Jake Son, Jon Clucas, Helen Xu, Bonhwang Koo, Anirudh Krishnakumar, Bharat Biswal, Francisco Castellanos, Stan Colcombe, Adriana Di Martino, Xi-Nian Zuo, Arno Klein
IntelliEppi: Intelligent reaction monitoring and holistic data management system for the molecular biology lab. Arthur Neuberger, Zeeshan Ahmed, Thomas Dandekar
Findings of a retrospective, controlled cohort study of the impact of a change in Nature journals’ editorial policy for life sciences research on the completeness of reporting study design and execution. Malcolm Robert Macleod, The NPQIP Collaborative group
A persistent lack of International representation on editorial boards in environmental biology. Johanna Espin, Sebastian Palmas-Perez,Farah Carrasco-Rueda, Kristina Riemer, Pablo Allen, Nathan Berkebile, Kirsten Hecht, Renita Kay Kastner-Wilcox, Mauricio Nunez-Regueiro, Candice Prince, Maria Constanza Rios-Marin, Erica P. Ross, Bhagatveer Sangha, Tia Tyler, Judit Ungvari-Martin, Mariana Villegas, Tara Cataldo, Emilio Bruna
Idea farming: it is a good idea to have bad ideas in science. Christopher J Lortie
Imagining the ‘open’ university: Sharing scholarship to improve research and education. Erin C McKiernan
The Modern Research Data Portal: A design pattern for networked, data-intensive science. Kyle Chard, Eli Dart, Ian Foster, David Shifflett, Steven Tuecke, Jason Williams
Can editors save peer review from peer reviewers? Rafael D’Andrea, James P O’Dwyer.
How to share data for collaboration. Shannon E Ellis, Jeffrey T Leek
An invitation to modeling: building a community with shared explicit practices. Kam D Dahlquist, Melissa L Aikens, Joseph T Dauer, Samuel S Donovan, Carrie Diaz Eaton, Hannah Callender Highlander, Kristin P Jenkins, John R Jungck, M Drew LaMar, Glenn Ledder, Robert L Mayes, Richard C Schugart
Down with ncRNA! Long live fRNA and jRNA! Dan Graur
What exactly is ‘N’ in cell culture and animal experiments? Stanley E Lazic, Charlie J Clarke-Williams, Marcus R Munafo
Estimating the scale of biomedical data generation using text mining. Gabriel Rosenfeld, Dawei Lin
Why not…
Visual art inspired by the collective feeding behavior of sand-bubbler crabs. Hendrik Richter
The reality of “food porn”: Larger brain responses to food-related cues than to erotic images predict cue-induced eating. Francesco Versace, David W Frank, Elise M Stevens, Menton M Deweese, Michele Guindani, Susan M Schembre


 (1 votes)
(1 votes)