3rd Crick Beddington Symposium: Anterior Fates Await Those Who Migrate – How Nodal Dynamics Shape Anteroposterior Patterning
Posted by Alex Neaverson, on 14 March 2025
Although I originally set out to highlight the work of early career researchers at the symposium, Professor Vasso Episkopou’s overwhelmingly enthusiastic response coupled with my interest in anteroposterior patterning made her an obvious choice for an interview. At the end of the first day, although I was tired from a day packed full of interesting presentations and engaging conversations, I approached her poster – which was an unconventional, yet effective, series of laminated PowerPoint slides!

Anteroposterior (AP) patterning in early vertebrate embryos is influenced by Nodal signalling – high levels promote anterior fates, while low levels drive posterior fates. You might therefore expect a gradient of Nodal along the primitive streak, but this is not the case; it is expressed ubiquitously in the streak. This discrepancy is the impetus of the research conducted by Vasso and her group at Imperial College London.
SnoN, a repressor of Nodal signalling, binds Smad4 to suppress anterior fate-promoting genes at Smad-binding elements (SBEs). Upon Nodal activation, phosphorylated Smad2/3 (pSmad) forms a complex with SnoN, which is recognized and degraded to its entirety by the ubiquitin ligase Arkadia (Rnf111). This surprising mechanism directly links pSmad levels to SnoN reduction, only forcing SnoN removal under high pSmad signaling. “The anterior target genes, they require Arkadia and high signalling, in order to degrade the repressor”, Vasso explained. The SBE is now clear, allowing pSmad-Smad4 to bind, and together with co-activators, initiate transcription of anterior genes. The necessity of Arkadia for anterior development is very clear in the headless phenotype of Arkadia-/- mice. In addition, head formation is rescued in Sno-/-;Ark-/- embryos, confirming that Arkadia is responsible for achieving the de-repression.
But Nodal signalling is abundant in the primitive streak – so how is AP patterning regulated? Vasso’s research group have identified that the dynamics of Nodal signalling control AP patterning in the primitive streak, rather than morphogen gradients. A TGF-B time course treatment of embryonic stem cells showed that, at high levels of signalling, there is a temporary decrease in SnoN at 1-2 hours due to its degradation by Arkadia, then SnoN levels increase again at 4-6 hours; this temporary reduction frees up space on the SBE for pSmad-Smad4 to activate transcription of anterior genes. The rising of SnoN levels at 4-6h is due to a negative feedback effect: one of pSmad’s early targets is SnoN itself, and the resulting increase in SnoN overwhelms Arkadia. This leads to a very transient activation of Arkadia-dependent anterior targets.
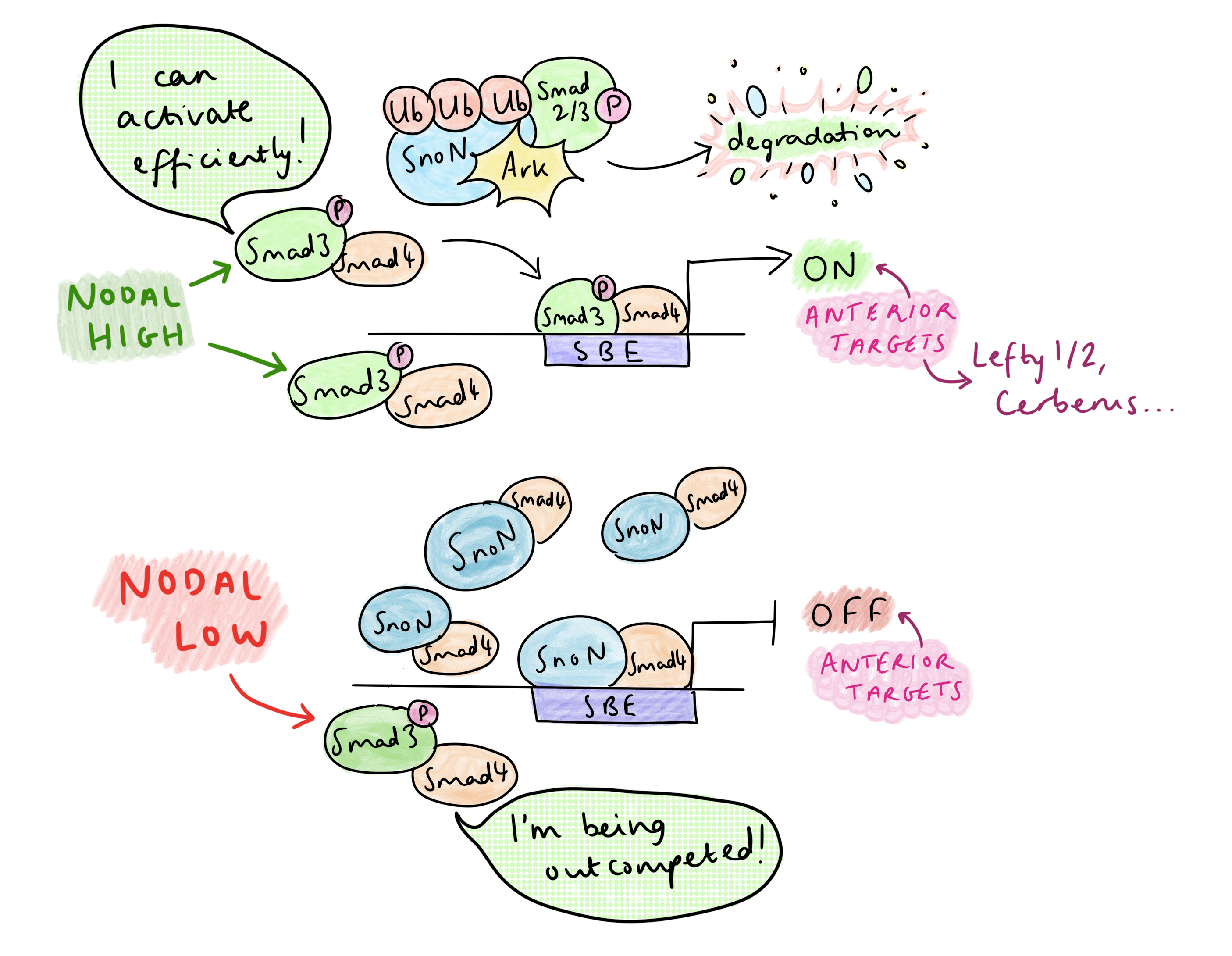
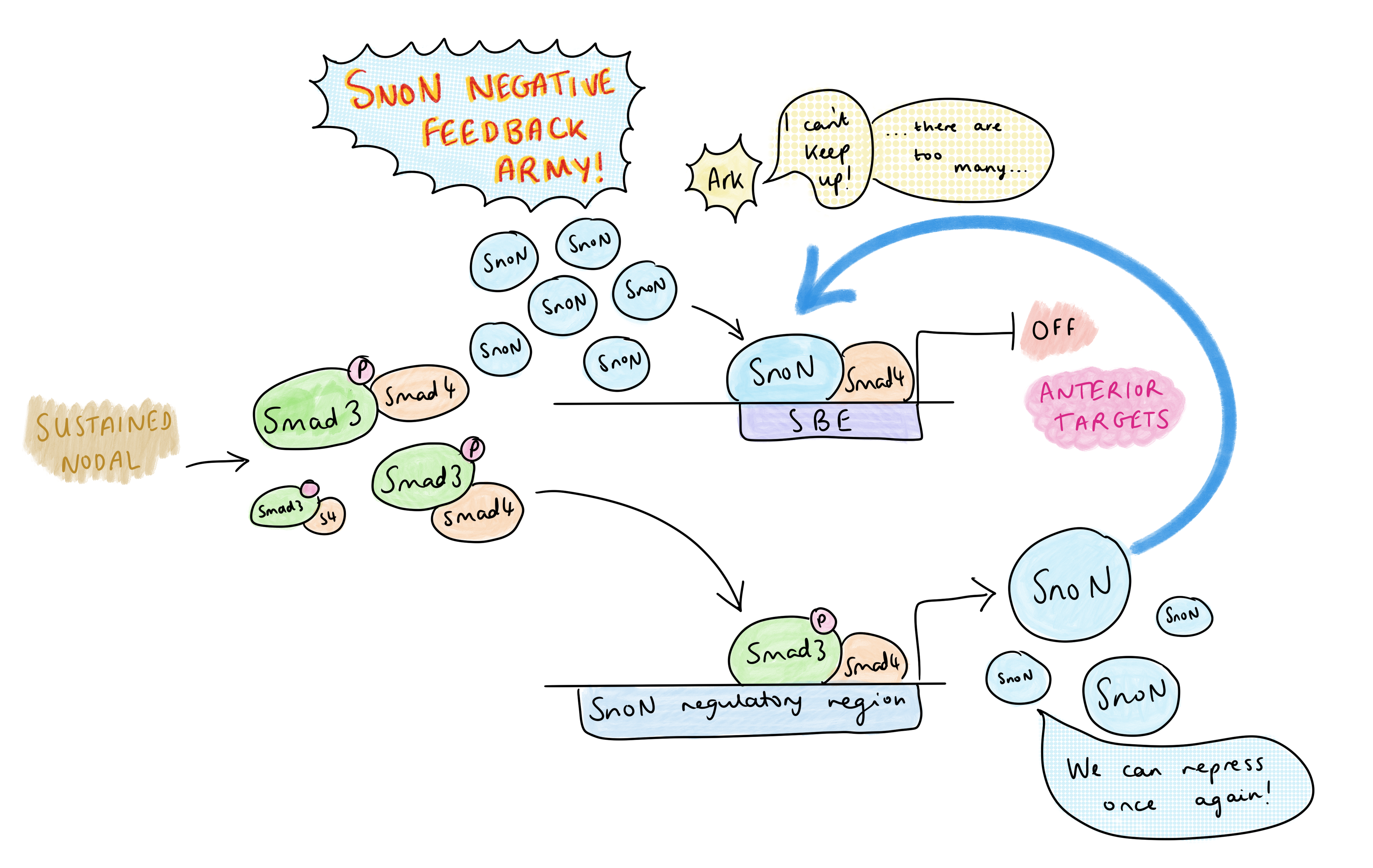
On the other hand, in the presence of lower levels of Nodal, SnoN is not degraded by Arkadia, so there is competition between SnoN-Smad4 and pSmad-Smad4 for binding the SBE. Posterior Nodal targets do not require the SBE to be cleared completely – only a low level of Nodal signalling is required to relax the chromatin by pausing histone deacetylation, which permits co-regulators of posterior targets to bind (at other sites) and activate transcription. Since posterior genes are co-regulated by other transcription factors, they only need Nodal signalling for partial de-repression to become activated. This contrasts with the anterior targets, which can only be activated by pSmad-Smad4 after clearing of the SnoN from the SBE.
Altogether, this means that anterior identity in the primitive streak must be acquired in a very short time window, in response to acute, high levels of Nodal signalling, before any negative feedback can kick in. Vasso has put this together with what we know about cell migration in the streak – the first cells to exit, the fastest migrating cells, leave the streak via anterior migration and become the anterior endoderm. These cells also express Nodal antagonists, such as Lefty 1/2 and Cerberus, doubling down to ensure that their exposure to Nodal is very short-lived. “Cerberus is a triple attack,” Vasso explained, “a Nodal, Wnt, and BMP inhibitor. So, these cells express antagonists to shield themselves from sustained (posteriorising) signals”. These antagonists activate the transcription of immediate early genes, ensuring that anterior identity is swiftly acquired. In contrast, slower migrating cells acquire posterior identity, since remaining in the streak for longer exposes them to sustained Nodal signalling, leading to the repression of anterior genes and allowing co-regulators to impose a posterior fate.
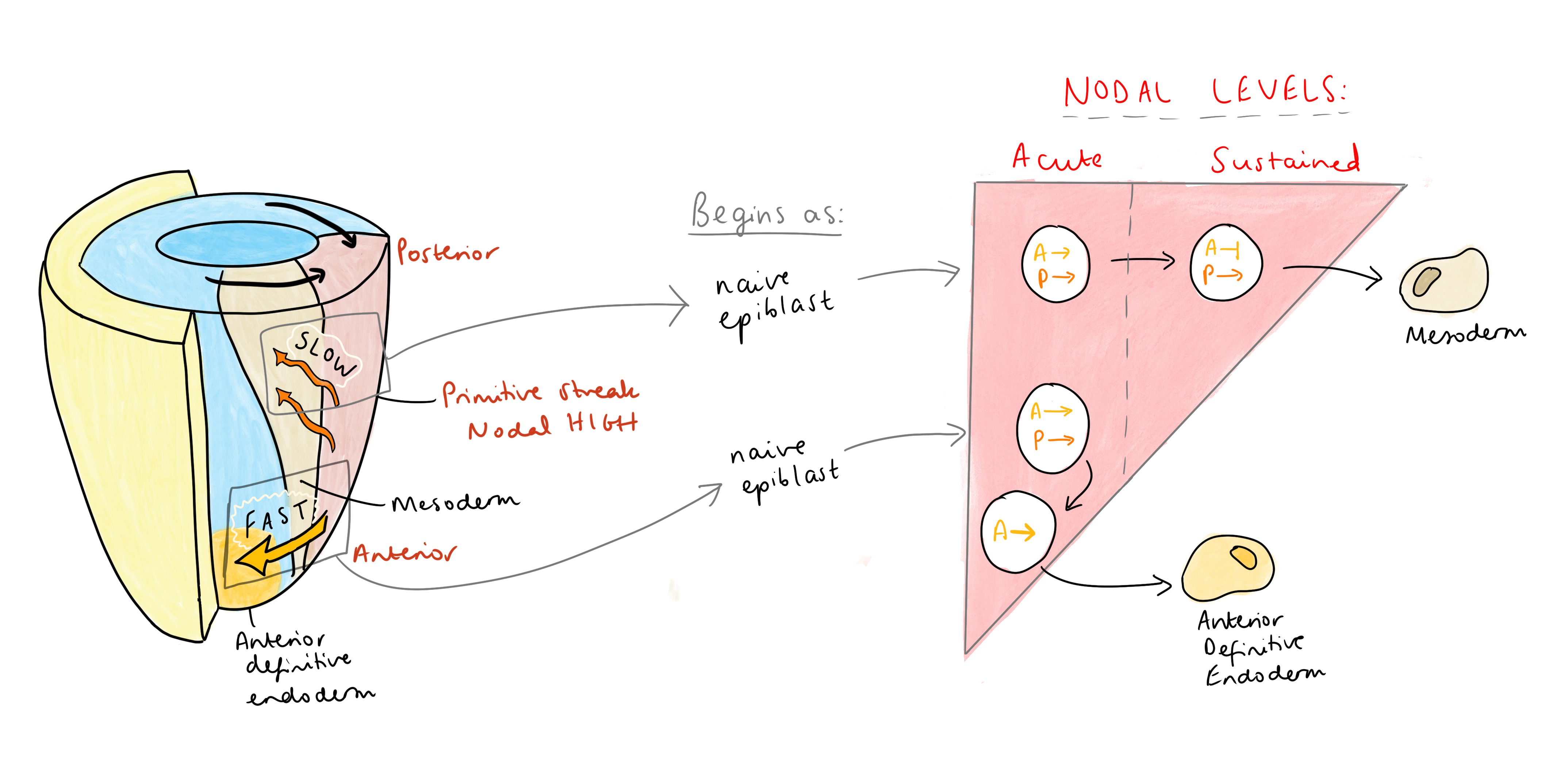
Vasso suggests that this dynamic regulation of signalling may be a widespread mechanism in fate determination. A similar principle applies to T lymphocyte differentiation: high TGFβ levels drive Treg differentiation in an Arkadia-dependent manner, whereas low levels drive Th17 differentiation independently of Arkadia (Xu et al., 2021). This model may extend to other biological contexts yet to be explored.
You can read Vasso’s group’s preprint on Nodal dynamics here:
Carthy, J.M., Ioannou, M. and Episkopou, V. (2019) ‘Arkadia via SNON enables NODAL-SMAD2/3 signaling effectors to transcribe different genes depending on their levels’. bioRxiv, p. 487371. Available at: https://doi.org/10.1101/487371.
Read about how the same system acts in T lymphocyte differentiation here:
Xu, H. et al. (2021) ‘Arkadia-SKI/SnoN signaling differentially regulates TGF-β–induced iTreg and Th17 cell differentiation’, Journal of Experimental Medicine, 218(11), p. e20210777. Available at: https://doi.org/10.1084/jem.20210777.
Stay tuned for more poster interviews coming soon!


 (2 votes)
(2 votes)