Novel mouse alleles allow for sequential mutagenesis using the dual recombinase technology
Posted by dkirsch, on 20 September 2015
Genetically engineered mouse models have been used extensively to study a wide variety of biological processes in vivo, and innovations in genetic engineering have made it possible to dissect more intricate biological questions. For example, the first mice that showed successful germline transmission of foreign DNA were created in the 1980s, and this allowed the generation of various transgenic “oncomice” such as the first mouse model of breast cancer using the MMTV-Myc transgene 1, and the first mouse model of osteosarcoma using the MT-FosLTR transgene 2. Subsequently in the early 1990s, mice were developed to have whole-body deletions of various genes such as the p53 tumor suppressor gene 3. The combination of the ability to introduce oncogenes and delete tumor suppressor genes through these initial transgenic mouse technologies contributed greatly to our current understanding of cancer development, progression, and treatment.
Nonetheless, transgenic mice and whole-body knock out mice sometimes do not survive development 4. To overcome this limitation, investigators harnessed site-specific recombinase systems from bacteriophages and yeasts such as the Cre-loxP and the Flp-FRT systems, which were first adapted to the mouse genome for germline transmission in the 1990s to enable temporally-regulated and tissue-specific genetic manipulations 5, 6. Since then, many conditional mouse alleles utilizing transgenic or endogenous promoter-driven Cre and loxP-regulated genes have been generated to increase tissue-specificity of gene expression and decrease pre-mature lethality and other unwanted phenotypes. Additionally, replication-defective viruses (adenoviruses, lentiviruses) containing Cre recombinase (Adeno-Cre, Lenti-Cre) have also been used to further improve upon the temporal regulation of gene expression 7. Further modifications enabled the viruses to carry specific promoter-driven Cre recombinases to add tissue specificity after viral delivery 8. Finally, similar temporal regulation can be achieved by using fusion proteins combining Cre and mutated hormone receptors such as the estrogen receptor (Cre-ERT2) 9. By utilizing this approach, metabolites of tamoxifen can be used to translocate Cre-ERT2 to the nucleus and thus activate subsequent Cre-mediated gene modification.
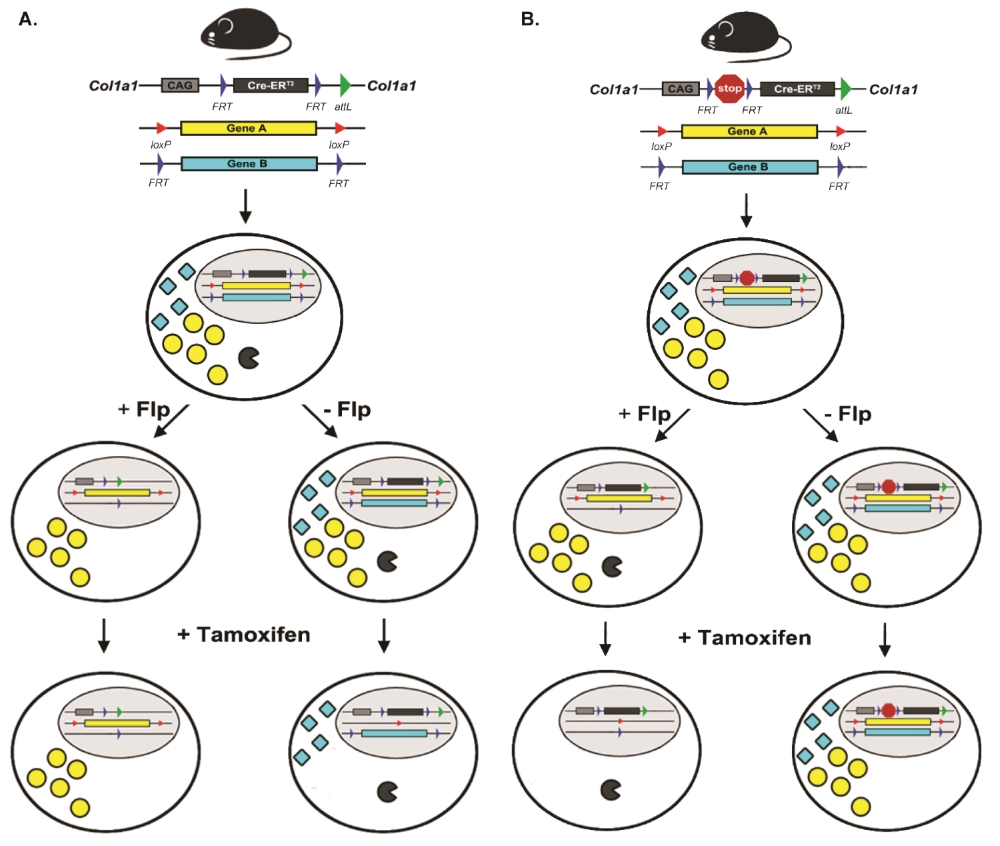
Recently, there has been increasing interest in combining more than one recombinase system in the same mouse model using dual recombinase technology to allow for sequential mutagenesis and to better model sporadic cancers in the adult mouse 10, 11. In our lab, we have generated two complimentary new mouse alleles that facilitates the regulation of Cre-ERT2 by the Flp-FRT recombinase system. Both alleles are knocked into the endogenous Col1a1 locus for ubiquitous expression, and also at the same time reserve the two Rosa26 alleles for other transgenes of interest. In the first mouse allele, Col1a1FRT-Cre-ER-T2-FRT, Cre-ERT2 is flanked by FRT sites. The rationale to generate Col1a1FRT-Cre-ER-T2-FRT mice is to enable whole animal ubiquitous expression of Cre-ERT2 until exposure to Flp recombinase (Figure 1A). After Flp-mediated recombination of the FRT sites, cells are no longer able to express Cre-ERT2 and therefore lose the ability to delete DNA flanked by loxP sites following exposure to tamoxifen. In this way, different mutations can be introduced in adjacent cells in vivo so that the consequences for intercellular interactions, such as cancer cells and stromal cells, can be studied. In the second mouse allele, Col1a1FRT-STOP-FRT-Cre-ER-T2, Cre-ERT2 sits downstream of a FRT-flanked STOP cassette, which inhibits transcription of Cre-ERT2. The rationale to generate Col1a1FRT-STOP-FRT-Cre-ER-T2 is that initially no cell expresses Cre-ERT2 because transcription of Cre-ERT2 is terminated by an upstream FRT site-flanked transcription STOP cassette (Figure 1B). However, after Flp-mediated recombination, the STOP cassette is excised. Therefore, these cells can initiate transcription of the Cre-ERT2 fusion protein, which in response to subsequent exposure to tamoxifen translocates into the nucleus to recombine DNA flanked by loxP sites. Cells without exposure to Flp will not be able to undergo Cre-mediated DNA recombination. In this way, the Col1a1FRT-STOP-FRT-Cre-ER-T2 allele enables sequential mutations within the same cell over time. First, one mutation occurs in the cell from Flp recombinase, and then tamoxifen activates Cre recombinase in the same cell to mutate a second gene to study how the order of gene mutations may affect cellular outcome. In addition, multiple genes may be mutated by Flp recombinase to initiate tumor development. Then, the role of a loxP-flanked gene in tumor maintenance can be studied because only the tumor cell will express Cre-ERT2. This allele can therefore be used to identify potential therapeutic targets.
Through characterization of the Col1a1FRT-Cre-ER-T2-FRT mice using the reporter allele Rosa26mTmG 12, we found that a single intraperitoneal injection of tamoxifen at 75mg/kg into Col1a1FRT-Cre-ER-T2-FRT; Rosa26mTmG/+ mice is sufficient to translocate the Cre-ERT2 fusion protein into the nucleus and mediate recombination of the loxP-sites flanking tdTomato. This resulted in the deletion of the tdTomato red fluorescent protein, and the subsequent expression of eGFP green fluorescent protein from cells of all tissues examined. The limitation of this model is that there is an age- and tissue-dependent Cre-ERT2 activation independent of tamoxifen administration, most notably in the pancreas and the liver. Therefore, in experiments involving these organs using the Col1a1FRT-Cre-ER-T2-FRT mice, age of the mice at the onset of Flp and tamoxifen administration is crucial for the interpretation of the results.
We also characterized the Col1a1FRT-STOP-FRT-Cre-ER-T2 mice using the reporter allele Rosa26mTmG. When Col1a1FRT-STOP-FRT-Cre-ER-T2; Rosa26mTmG/+ mice were given a single intraperitoneal injection of tamoxifen at 75mg/kg, there was no Cre-ERT2-mediated deletion of the tdTomato red fluorescent protein or expression of the eGFP green fluorescent protein. Next, we generated a mouse model of soft tissue sarcoma using the new allele to verify the ability of the STOP cassette to be removed by Flp, and the subsequent ability of Cre-ERT2 to manipulate loxP-flanked alleles. In this model, Col1a1FRT-STOP-FRT-Cre-ER-T2; KrasFRT-STOP-FRT/+; p53FRT/FRT; Rosa26mTmG/+ mice were first administered intramuscular injection of adenovirus carrying Flp recombinase in the hindlimb to form soft tissue sarcomas at the site of injection via the expression of the oncogenic KRAS protein and elimination of the p53 tumor suppressor protein 11, 13, 14. Following the formation of soft tissue sarcomas, either single intratumoral injection of 4-hydroxytamoxifen or several doses of systemic 4-hydroxytamoxifen were administered. The resulting tumor showed tumor parenchymal cells with deletion of tdTomato red fluorescent protein and expression of eGFP green fluorescent protein, while the tumor stromal cells such as the vasculature continued to express tdTomato. One potential challenge in this model is that while the Col1a1FRT-STOP-FRT-Cre-ER-T2 is functional, single intraperitoneal injection of tamoxifen failed to activate Cre-ERT2 in primary sarcomas. Therefore, the delivery of an adequate level of tamoxifen and/or its metabolites into the tumor is critical to activate Cre-ERT2 expression with this allele.
We anticipate that these two new alleles will allow for increased control of gene manipulation in genetically engineered mouse models. These alleles, in conjunction with other new technologies in the field such as the CRISPR/Cas9 system 15, have the potential to bring together the speed and efficiency of the CRISPR/Cas9 system with the spatial and temporal control of dual recombinase technology, manipulate the genome in vivo to study development, cancer and other diseases in the mouse.
Minsi Zhang and David Kirsch, Department of Radiation Oncology, Duke University Medical Center
Main reference
Zhang, M., & Kirsch, D. The generation and characterization of novel Col1a1FRT-Cre-ER-T2-FRT and Col1a1FRT-STOP-FRT-Cre-ER-T2 mice for sequential mutagenesis. Disease Models & Mechanisms. 2015. 8(9), 1155-1166 DOI: 10.1242/dmm.021204
Other references:
- Stewart TA, Pattengale PK, Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38(3):627-37. PubMed PMID: 6488314.
- Ruther U, Komitowski D, Schubert FR, Wagner EF. c-fos expression induces bone tumors in transgenic mice. Oncogene. 1989;4(7):861-5. PubMed PMID: 2547184.
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr., Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215-21. doi: 10.1038/356215a0. PubMed PMID: 1552940.
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359(6393):295-300. doi: 10.1038/359295a0. PubMed PMID: 1406933.
- Orban PC, Chui D, Marth JD. Tissue- and site-specific DNA recombination in transgenic mice. Proc Natl Acad Sci U S A. 1992;89(15):6861-5. PubMed PMID: 1495975; PubMed Central PMCID: PMC49604.
- Dymecki SM. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc Natl Acad Sci U S A. 1996;93(12):6191-6. PubMed PMID: 8650242; PubMed Central PMCID: PMC39212.
- Wang Y, Krushel LA, Edelman GM. Targeted DNA recombination in vivo using an adenovirus carrying the cre recombinase gene. Proc Natl Acad Sci U S A. 1996;93(9):3932-6. PubMed PMID: 8632992; PubMed Central PMCID: PMC39462.
- Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19(6):754-64. doi: 10.1016/j.ccr.2011.04.019. PubMed PMID: 21665149.
- Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27(22):4324-7. PubMed PMID: 10536138; PubMed Central PMCID: PMCPMC148712.
- Shai A, Dankort D, Juan J, Green S, McMahon M. TP53 Silencing Bypasses Growth Arrest of BRAFV600E-Induced Lung Tumor Cells In a Two-Switch Model of Lung Tumorigenesis. Cancer research. 2015. doi: 10.1158/0008-5472.CAN-14-3701. PubMed PMID: 26001956.
- Schonhuber N, Seidler B, Schuck K, Veltkamp C, Schachtler C, Zukowska M, Eser S, Feyerabend TB, Paul MC, Eser P, Klein S, Lowy AM, Banerjee R, Yang F, Lee CL, Moding EJ, Kirsch DG, Scheideler A, Alessi DR, Varela I, Bradley A, Kind A, Schnieke AE, Rodewald HR, Rad R, Schmid RM, Schneider G, Saur D. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat Med. 2014;20(11):1340-7. doi: 10.1038/nm.3646. PubMed PMID: 25326799; PubMed Central PMCID: PMC4270133.
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593-605. doi: 10.1002/dvg.20335. PubMed PMID: 17868096.
- Moding EJ, Lee CL, Castle KD, Oh P, Mao L, Zha S, Min HD, Ma Y, Das S, Kirsch DG. Atm deletion with dual recombinase technology preferentially radiosensitizes tumor endothelium. J Clin Invest. 2014;124(8):3325-38. doi: 10.1172/JCI73932. PubMed PMID: 25036710; PubMed Central PMCID: PMC4109553.
- Moding EJ, Castle KD, Perez BA, Oh P, Min HD, Norris H, Ma Y, Cardona DM, Lee CL, Kirsch DG. Tumor cells, but not endothelial cells, mediate eradication of primary sarcomas by stereotactic body radiation therapy. Sci Transl Med. 2015;7(278):278ra34. doi: 10.1126/scitranslmed.aaa4214. PubMed PMID: 25761890; PubMed Central PMCID: PMC4360135.
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440-55. doi: 10.1016/j.cell.2014.09.014. PubMed PMID: 25263330; PubMed Central PMCID: PMC4265475.


 (1 votes)
(1 votes)