Forgotten classics: Rosa Beddington’s chimeras
Posted by the Node, on 11 November 2016
R. S. P. Beddington (1981). An autoradiographic analysis of the potency of embryonic ectoderm in the 8th day postimplantation mouse embryo. Journal of Experimental Embryology and Morphology. 64: 87-10. Open Access
R. S. P. Beddington (1982). An autoradiographic analysis of tissue potency in different regions of the embryonic ectoderm during gastrulation in the mouse. Journal of Experimental Embryology and Morphology. 69: 265-285. Open Access
Recommended by Patrick Tam, University of Sydney
The original chimera was a beast made out of different beasts, with “the head of a lion and the tail of a serpent, while her body was that of a goat, and she breathed forth flames of fire”. Today’s biomedical definition is not too far from Homer’s, a chimera being an organism made up of cells from more than one zygote. As an experimental technique, chimeras have been used for decades to define the developmental potential of cells and the influence of the environment cells find themselves in (does a cell stick to its original fate if transplanted to a different location? Does the environment override an initial choice?). Even today, one stringent test for pluripotency is the ability of transplanted cells to give rise to all cell types in the chimeric organism.
The first mouse chimeras (1, 2) were made by aggregating two pre-implantation (cleavage stage) embryos together: the resultant embryos, twice the normal size, gave rise to normal sized chimeric pups. Subsequent techniques involved injecting exogenous cells into blastocysts. But what about the decisions that happen in later stages? Post-implantation mouse chimeras were pioneered by Rosa Beddington, and her two single author papers describing the work, published in 1981 and 1982 in the Journal of Embryology and Experimental Morphology (the forerunner of Development), are the subject of this Forgotten Classics highlight.
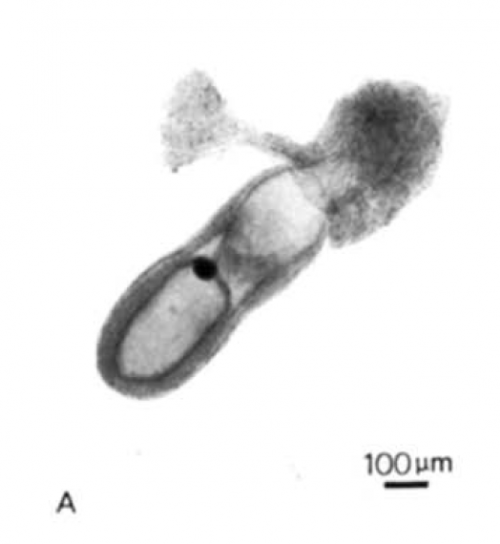
The two papers should be read together, the second being a continuation and expansion of the first. The question that drove the project concerned the patterning of the epiblast (which was then referred to as the embryonic ectoderm), the tissue that gives rise to the embryo proper. The problem was articulated by Beddington as follows:
“During gastrulation the single epithelial sheet of embryonic ectoderm is converted into a highly complicated form, made up of a variety of tissue types and embodying the basic design of the foetus. This means that the key to foetal organization must lie in the orderly allocation of tissue primordia within the embryonic ectoderm”
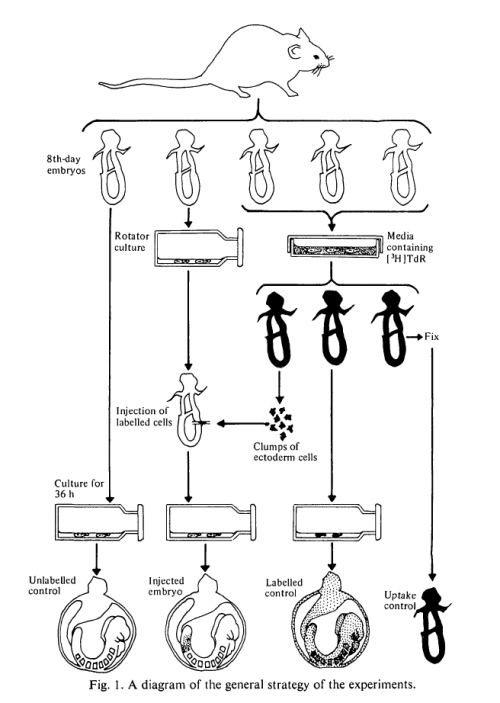
The problem was the inaccessibility of the mouse embryo for experimental manipulation. In Beddington’s time, while rats could be cultured ex utero until the end of gastrulation, mouse culturing techniques did not achieve comparable successes. Her papers describe a technique by which embryos are dissected from the uterus at day 8, at the late-primitive-streak stage, and then cultured for 36 hours in rat serum. These 36 hours are “a time of intensive cell division and differentiation and also marked by substantial morphogenetic activity”. The culture produced early-somite-stage embryos that look normal when compared to in utero counterparts, with some minor differences (they were more translucent, and had expanded yolk sacs). The work was a technical feat: dissecting early mouse embryos is not easy, yet Beddington was blessed with legendary dissecting skills.
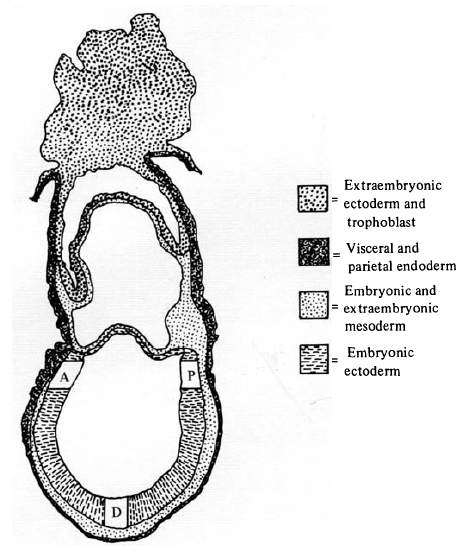
This technique allowed the assessment of epiblast cell lineage in the intact embryo. As encountered in the last Forgotten Classic, cell lineage analysis requires a marker, and Beddington chose 3H-thymidine (3H-T), a radiolabelled nucleoside you can visualise with autoradiography. In many ways 3H-T is not the ideal marker: unlike genetic markers it dilutes with cell divisions; subsequent work indicated it can inhibit DNA synthesis and be cytotoxic (although this does not seem to have been a problem with Beddington’s work); and, perhaps most annoyingly, once you have your stained and sectioned embryos, you have to cover them with autoradiographic stripping film for three weeks before processing and analysing the film. Three weeks! And even then, after this intricate and demanding procedure, defective processing meant Beddington had to discard whole batches of slides.
The method was to bathe day 8 embryos in 3H-T, remove cells from different regions of the epiblast, inject them into unstained, synchronous hosts, and see where the labelled cells ended up after 36 hours in culture. An elegant aspect of the work is the controls: controls that hadn’t been labelled; controls that had been bathed in 3H-T and cultured without dissection; controls that had been fixed before culturing. All of these experiments, diagrammed with characteristic artistry in the figure, were also carried out with reference to in utero development.
With the method established – labelled controls looked pretty much the same as unlabelled controls, and the labelled cells could colonise host tissues and did not form structures you would not expect to see – the stage was set to address her main questions. Was cell fate spatially patterned in the epiblast? In other words, could you draw a fate map? And how plastic was this fate?
Beddington performed two types of injection. Orthotopic injections involved like-for-like injections, with labelled epiblast from a particular donor region injected into the same region in the host. These injections showed that different regions of the epiblast gave rise to different parts of the post-gastrulation embryo: for instance, distal epiblast could contribute to somites and notochord, but anterior epiblast could not. As Beddington acknowledged, this may not have been particularly surprising given results in the chick, but it was important to demonstrate that there was a consistent regionalised pattern of tissue allocation during gastrulation. You could begin to map the post-gastrulation embryo back to the epiblast.
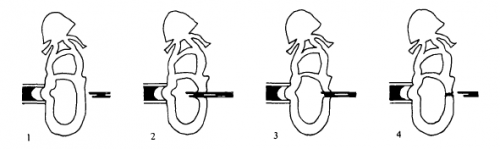
What orthotopic transplantation cannot reveal is whether this regionalisation reflects an inherent cell fate, or the consequence of epiblast cells perceiving extrinsic cues. Heterotopic transplantation, putting donor cells in a location in the host that is different from where they came from, allowed Beddington to get at this. She found no evidence for rigid cell fate in the epiblast: when transplanted to a different location, cells readily contributed structures other than those they form normally (although there were some ‘propensities’ of certain cells to contribute to one structure or another, suggesting some degree of cell fate restriction). Thus, cell fate at the epiblast stage is ‘plastic’, and could be readily influenced by the environment the cells find themselves in. So the story is a mix: you can draw a fate map on the epiblast, but the cells are happy, if we labour the metaphor a bit, to learn a new language when transplanted into another country.
These papers represent a landmark of mouse embryology. In the following decades, many of the molecular players in epiblast patterning have been identified, as well as the mechanisms that maintain epiblast cell potency, supporting and expanding Beddington’s work. The recent BSDB meeting celebrating the present and future of chimeric research shows that chimeras still have as much to tell us about development as they taught Rosa Beddington in the early 1980s.
Thoughts from the field
Patrick Tam, University of Sydney
“The findings of these works have revealed the regionalisation of cell fates in the germ layers of the gastrulating mouse embryo, pointing to the establishment of a basic body plan. These studies also outlined an experimental paradigm for the analysis of cell fate and potency in a mammalian embryo using innovative techniques of micromanipulation, lineage tracking and whole mouse embryo culture.
Recently, there is heightened interest in the application of these techniques to generate post-implantation chimeras for assessing the differentiation potential of stem cells, such as mouse epiblast stem cells and human pluripotent stem cells1 . It is therefore timely and newsworthy to highlight Rosa’s papers.”
1: Tam PPL (2016) Human stem cells can differentiate in post-implantation mouse embryos. Cell Stem Cell 18: 3-4. PMID 26748747 DOI:10.1016/j.stem.2015.12.010
Virginia Papaioannou, Columbia University Medical Center
“It is hard to remember a time before the fate map of the mammalian embryo had experimental backing, but in the late 70s, as mammalian embryo culture and manipulation techniques were just beginning to make great strides, it was the chick fate map that guided us and provided a template for investigation. These two papers by Rosa Beddington were remarkable in adapting newly devised rat embryo culture methods to allow investigation of mouse postimplantation embryos ex utero and, for the first time, developing methods for making experimental postimplantation mouse chimeras. Rosa used these techniques to approach two related aspects of embryonic cells: fate and potential, fate being the normal differentiation outcome of a cell in undisturbed development and potential being what that cell is capable of doing in altered circumstances, such as being placed heterotopically in a different position in the embryo.
These two papers comprise the bulk of Rosa’s thesis work for her D.Phil. from Oxford University in 1981. I was privileged to serve as her supervisor for this work and rereading the papers brought back the sheer gutsiness of this brilliant young student as she pioneered a new methodology for mouse embryology. It also sent me searching for my copy of her dissertation, typewritten, with original photos taped onto the pages, where I marvelled anew at the camera lucida drawings of serial sections with labelled cells marked in pen, and hand drawn embryos with color-coded fate maps in coloured pencil.
In the thesis, the work was divided into a chapter on cell fate (orthotopic tissue transplants in synchronous embryos) and a separate chapter on cell potency (heterotopic transplants in synchronous embryos). In the subsequent publications this fate vs. potential distinction is not highlighted and both papers refer to “potency” in the title. Although I don’t remember why this was done, I imagine it was because of Rosa’s exactitude in the definition of the terms, as even synchronous orthotopic tissue transplantation is a disruption and might be subtly altering ‘normal’ cell fate. As we understand more about altering cell states in the age of induced pluripotent stem cells, this assiduous attention to terminology and methodological detail is more relevant than ever. Rosa’s papers, nonetheless, ushered in an era of rapid advances in understanding cell fate and potential in the postimplantation mammalian embryo.”
Aidan Maartens
This post is part of a series on forgotten classics of developmental biology. You can read the introduction to the series here and read other posts in this series here. We also welcome suggestions for future Forgotten Classics.


 (8 votes)
(8 votes)