BSDB Gurdon Summer Studentship Report (15)
Posted by BSDB, on 1 November 2017
![]() Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding.
Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding.
Our fourth report from the 2017 group of student awardees comes from Eleanor Sheekey (student at University of Cambridge), who undertook her studentship with Peter Rugg-Gunn at the Babraham Institute in Cambridge.
Much ado about Nanog
 Named after the mythical “Land of the young”, Tír na nÓg, Nanog is a homeobox transcription factor expressed in embryonic stem cells (ESCs) aiding continual cellular proliferation alongside the other main pluripotency factors, Oct4 and Sox2. However, despite its contribution to pluripotency, much is still unknown of the mechanistic roles that Nanog plays within stem cells, since cells kept in culture deficient in Nanog retain the ability to self-renew (1). Interestingly, it was recently found that Nanog provides a link between pluripotency and chromatin organisation suggesting important functions beyond transcriptional regulation (3). With the BSDB Gurdon Studentship, I was privileged to spend my 8 weeks at the Babraham Institute alongside Dr Clara Novo to continue to explore this link.
Named after the mythical “Land of the young”, Tír na nÓg, Nanog is a homeobox transcription factor expressed in embryonic stem cells (ESCs) aiding continual cellular proliferation alongside the other main pluripotency factors, Oct4 and Sox2. However, despite its contribution to pluripotency, much is still unknown of the mechanistic roles that Nanog plays within stem cells, since cells kept in culture deficient in Nanog retain the ability to self-renew (1). Interestingly, it was recently found that Nanog provides a link between pluripotency and chromatin organisation suggesting important functions beyond transcriptional regulation (3). With the BSDB Gurdon Studentship, I was privileged to spend my 8 weeks at the Babraham Institute alongside Dr Clara Novo to continue to explore this link.
Centromeres and chromatin
A key hallmark of stem cells is their ability to divide. However, this division must be executed precisely to avoid aneuploidy, an abnormal chromosomal number in a cell, and prevent unregulated division and cancer formation. Centromeres play a crucial role in chromosome separation, composing centric heterochromatin (CH) for kinetochore formation and pericentromeric heterochromatin (PCH) to hold chromatids together (2). Although centromeres are thought to be defined epigenetically, the centro- and pericentromere contain minor and major satellite DNA repeats, respectively. Nanog binds the major satellite DNA repeats within the pericentromere, maintaining the PCH in a ‘more’ open state with increased transcription of major satellites and lower levels of H3K9me3(3), bridging the gap between pluripotency and chromatin organisation. It also promotes the formation of chromocenters, which are clusters of major satellite repeats from several chromosomes. Deletion of Nanog leads to chromatin compaction including the PCH. Since PCH is essential for genetic stability and Nanog levels are known to fluctuate in stem cell cultures, the consequences of an altered PCH organisation will need to be understood before successfully developing more stably reprogrammed stem cells for future medical treatments (4).
Meet E14 and BT12
Throughout my project, I was working with mouse ESCs (mESCs). Much like in a vending machine, each of the different genetically modified mESCs are assigned a systematic name. E14 is like salted crisps, a male, wild type mESC cell line, originally isolated from the inner cell mass of a developing embryo. BT12, on the other hand is spiced up, no longer containing Nanog, but instead expressing a GFP transgene (1). Maintaining these two cell lines in culture was essential for conducting my experiments to study the effect of Nanog-/-.
FISH, IF and ChIPs
As Nanog was shown to affect heterochromatin at pericentromeres, we wondered if this effect extended to the centromere. To tease apart any differences in protein levels and localisation at the centromeres between E14 and BT12, we used a combination of techniques. Immunofluorescence (IF) provides a clear single cell insight and is beautiful to visualise under the microscope. Fluorophores had to be carefully chosen to avoid the wavelength clashing with that of GFP (already being expressed in BT12) and to differ from each other. Fluorescence in situ hybridisation (FISH) visualises specific locations of DNA using single-stranded probes and thus in combination with IF provides a good indication of whether proteins localise to a particular DNA region.
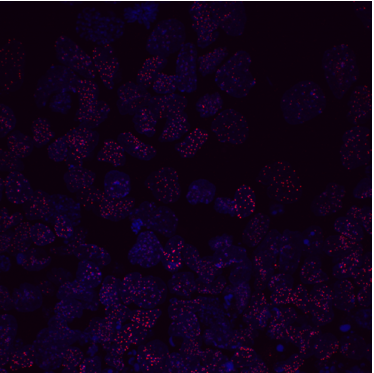

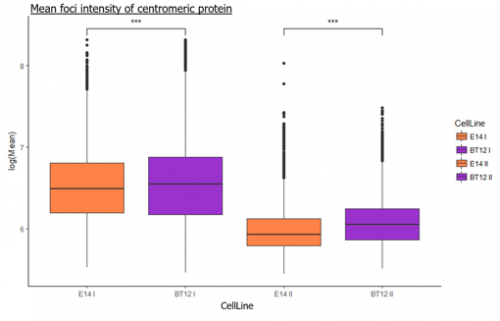
We were able to take some stunning images (Figure1) which could be used to quantify the levels of centromeric proteins assayed in wild-type and Nanog knockout cells. We then used the Imaris software (5) to quantify signals obtained from multiple 3D stacks, allowing for easy identification of fluorescent foci (Figure 2). Once the quantification of protein intensity was gathered I analysed it on R-studio to test for significant differences between both cell types (Figure 3).
To further this analysis, we used Chromatin immunoprecipitation (ChIP) that can answer many biological questions associated with DNA-protein interactions by fixing proteins to chromatin. In brief, by using antibodies specific to the protein of interest and magnetic beads that bind to these antibodies, a ‘pull-down’ of captured DNA can be isolated. The eluted DNA can then be analysed using ChIP-quantitative polymerase chain reaction (ChIP-qPCR) to quantify protein intensity at specific DNA regions. In our case, by using primers complementary to the major and minor satellites, we could assess if the binding of our proteins of interest differed between the cell lines.
Epilogue
Much like predicting the murderer in an Agatha Christie novel, one of my favourite aspects of the project was piecing together the experimental evidence to suggest hypotheses to explain our results. It is quite feasible for the absence of Nanog to have a diverse range of effects on the mESCs due to its interactions with DNA and other pluripotency factors, which could result in mislocalisation and/or altered post-translational modifications of its partners. Nevertheless, my preliminary results will require further validation and follow up before any conclusions can be made. However, in the manner of Shakespeare’s comedy, it has definitely been a fun experience, one from which I have gained much more lab confidence and exposure from, of which I am grateful both to the Rugg-Gunn lab at the Babraham Institute and the BSDB studentship that helped fund it.

References:
- Chambers et. al. Nanog safeguards pluripotency and mediates germline development Nature 450, 1230-1234 (2007)
- Probst, A. V. and Almouzni, G. (2008), Pericentric heterochromatin: dynamic organization during early development in mammals.Differentiation, 76: 15–23
- Novo CL, Tang C, Ahmed K, et al. The pluripotency factor Nanog regulates pericentromeric heterochromatin organization in mouse embryonic stem cells.Genes & Development. 2016
- Lopes Novo, C., & Rugg-Gunn, P. (2016). Crosstalk between pluripotency factors and higher-order chromatin organization.Nucleus, 7(5), 447–452.
- http://www.bitplane.com/imaris/imaris


 (3 votes)
(3 votes)