A journey towards uncovering the mechanics of embryonic polarization
Posted by Shyi-Chyi, on 17 February 2018
The key results of our recent paper in Nature Cell Biology
Cell polarization defines the spatial biological specificities in a cell. During the first cell cycle of a C. elegans zygote, its symmetry is broken by local remodeling of the cortical actomyosin network. This leads to a segregation of the dedicated polarity regulators, the PAR proteins, into two discrete cortical domains. However, it remains unclear how the mechanical changes driven by actomyosin contractions are transmitted to PAR proteins, and this regulates cellular spatial patterning. In our paper, we revealed that both actomyosin contractions and CDC-42 activity modulate the dynamics of the anterior PAR proteins (aPARs) through stimulation of their clustering at the cell cortex. In the early phase of polarity establishment, contractility of the actomyosin cytoskeleton drives cortical tension, which promotes clustering of PAR-3 (Mov.1, Fig.1). In turn, PAR-3 clusters recruit PKC-3, whereas CDC-42 activity antagonizes PKC-3 clustering during later phase of polarization (Mov.2, Fig.2). The degree of aPARs clustering is significantly associated with the stagnation of aPARs exchange at the cortex, and an effective entrainment with the advective flows of the cortical actomyosin network. This is the first report showing that the cell polarity protein PAR-3 is mechanosensitive, and senses forces to form higher order structures at the cell cortex. These findings depict how actomyosin and PAR proteins interplay and set up asymmetry (Fig.3). This feature may also play a role in other cell patterning events including neuron maturation, wound healing and cancer formation.
Movie 1: Cortical PAR-3::GFP (green) and NMY-2::mKate2 (magenta) during the first cell cycle of a C. elegans embryo.
How I started this project
I had made up my mind to become a researcher during my undergraduate days, after first trying to do a simple plasmid construction in a biological laboratory. Back then I was fascinated by all the equipment and materials in the lab. I went on to obtain my Master’s and Ph.D. degrees in Virology and Neuroscience respectively, before moving to Singapore to commence my first postdoctoral training in Frederic Bard’s lab at the Institute of Molecular and Cell Biology (IMCB), A*STAR. There, I matured as a scientist, and with access to multiple advanced microscopes across the various institutes within the campus, I was able to develop my working skills and knowledge in live-imaging techniques. These abilities, have turned out to be a great help in my career so far.
Before joining Fumio Motegi’s lab at the Temasek Lifesciences Laboratory (TLL), I had worked for over 10 years using cell culture to address different biological questions. So, I had a strong feeling that I must do something different for my next post-doc training. However, it was only after Fumio showed a fantastic movie depicting how anterior and posterior PARs are segregated in worm embryos that I decided to pursue this new area of study. My interests had been captured by how complex, yet exquisite, the underlying regulations can be. After I started working on this project and got some beautiful images of the genome-edited GFP-tagged aPARs, which were generous gifts from Kenneth Kemphus from the Cornell University, I realized that C. elegans is such a powerful tool with which we can address fundamental and important questions. I was also encouraged by the worm society, who kindly shared their materials and information, and welcomed me as a newcomer in their community. I was confident that this was what I wanted to do.
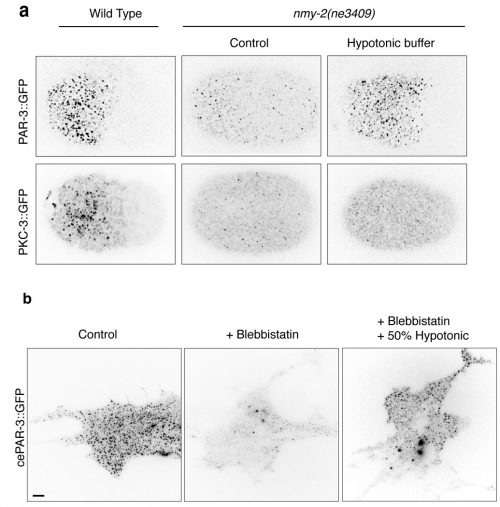
How I found the key results in this paper
The first Eureka moment came when I was inspecting the cortical PAR-3::GFP and PKC-3::GFP in wild-type embryos. They showed beautiful and dynamic puncta structures. This led us to further characterize the implication of those puncta during polarity establishment. We then conducted intensive examinations on the clustering of aPARs in wild-type and polarity-mutant zygotes. Another moment of insight came to me when my colleague Ms. Tricia Low noticed that PAR-3::GFP may exhibit mechanosensitivity. PAR-3::GFP failed to form clusters in the non-muscle myosin-II (nmy-2) mutant embryos, suggesting actomyosin and PAR-3 clustering were somehow connected. The clues became clearer when we observed another two phenomena. Firstly, in wild-type embryos, PAR-3 did not reside with NMY-2, suggesting that PAR-3 clustering is not caused by the direct recruitment of PAR-3 molecules to actomyosin. Secondly, although PAR-3 was unable to form clusters in nmy-2 zygotes, PAR-3 clustering was restored when the embryos underwent shape changes, with cortical ruffling occurring during the later stage of embryogenesis. This suggested that actomyosin contractility itself was not essential for PAR-3 clustering. Instead, physical properties of the cortex appeared to play a crucial role in the control of PAR-3. Hence, we hypothesized that cortical tension driven by actomyosin could promote PAR-3 clustering. We tested and validated this assumption in worm zygotes, as well as mammalian fibroblasts NIH3T3 cells exogenously expressing C. elegans PAR-3, which is achieved with the fantastic help from Dr. Yukako Nishimura from the Mechanobiology Institute (MBI), Singapore. We found that in both cases, PAR-3 clustering was rescued under hypotonic treatment when myosin wasn’t functional. This is the first report stating that PAR-3 reorganizes to higher-order structures in response to tension!
Movie 2: GFP tagged PAR-3, PKC-3, and CDC-42 during polarity establishment in one-cell C. elegans embryos.
How we build a team to test our hypothetical model
When we discovered that PAR-3 carries mechanosensitivity and that it could sense and respond to changes in tension in worm zygotes, we wanted to validate and replicate this experiment in cultured cells and monitor PAR-3 clustering using a TIRF microscope. In the meantime, we also needed a proper method to quantify and demonstrate the clustering so that we could report its significance under different conditions. To allow the research to progress without delay, we decided to collaborate with researchers outside of our lab. The MBI and the IMCB instantly came to mind. At MBI, there are dozens of experts in the field of physical biology, as well as a comprehensive collection of advanced microscopes. We therefore felt there was no better choice for collaboration. Moreover, I had established tight connections with people in the IMCB during my post-doctoral training there. In particular, with the Computational Bioimage Analysis (CBA) Unit, which is run by truly reliable and efficient staffs who are doing great jobs in image analysis and quantification. I was fortunate to have the opportunity to establish fruitful collaborations across TLL, MBI, and IMCB. Each institute is highly-respected not only in Singapore, but worldwide, for consistently publishing in high-profile journals, and attracting talented scientists from different countries. Singapore had become one of the most splendid research hubs in the world, which is attributed to the massive investment and strong support from the government into the life science realm.
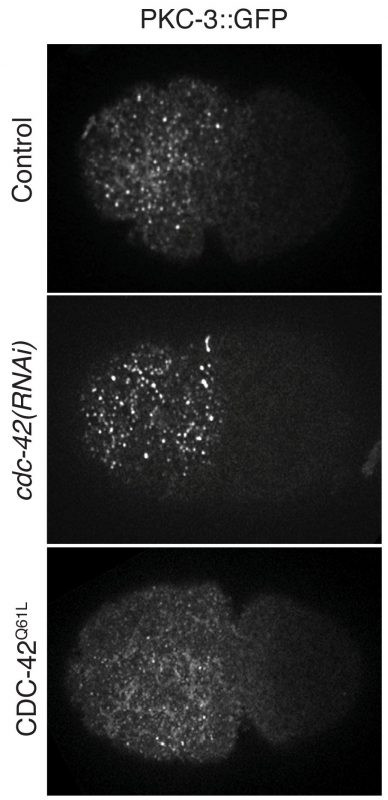
How we struggled to publish our project
I think the most difficult time for us along the journey was when our manuscript was rejected by the journal in December of 2016. The reviewers appreciated the main point of our story, but they thought the quantification method we had used to measure aPARs clustering was insufficient. Time was a big concern as we considered whether to submit to another journal, which might have been the easier path to publication, or stick with the same journal and strengthen our manuscript. We were in a dilemma. But since we were confident in ourselves, and in our story, we immediately made the decision to amend the quantification method and reanalyze all of our images. Fortunately, the results returned by the modified methods were consistent with our previous conclusions. We quickly revised the manuscript and resubmitted to the same journal and it turned out to be a happy ending. We appreciate Dr. Weimiao Yu and Dr. Laurent Gole from the Computational Bioimage Analysis (CBA) Unit of IMCB for their efforts to come up with the modified code, as well as the detailed descriptions for the quantification method, for the revised manuscript. We are also very grateful to the editor and all our reviewers for their constructive comments and positive encouragements to improve our manuscript. We did enjoy the review process. I hope what we had gone through can be an inspiration to those who might currently be struggling in a difficult time like this. What we need to do is to keep calm and believe in yourself.
Publication of our work along with two other groups
We learned there were several groups working on a similar topic since 2016. Fortunately, we maintained unencumbered communication with these groups, through emails or in person during international conferences, and it is truly exciting to see that we have all successfully published our stories in high-impact journals.
The common conclusion drawn by the three groups is that the degree of PAR-3 oligomerization increases during polarity establishment, and this further promotes the formation of aPAR clusters. The clustering of aPARs supports its segregation along with the actomyosin flow. Moreover, single-molecule pull-down in single embryos, as well as particle tracking, were used by Dickinson et al. to demonstrate that clustered PAR-3 is coupled to the cortical flow and phosphorylation by the mitotic kinase PLK-1 prevents PAR-3 from undergoing oligomerization. Furthermore, through a functional assay, Rodriguez et al. showed that PKC-3 inhibition or activation depends on its interaction with pools of PAR-3 or CDC-42, respectively. We found that PAR-3 carries mechanosensitivity that responds to cortical tension driven by actomyosin contraction. It’s genuinely fantastic that all three papers support each other and yet, by employing distinct approaches, present a unique significance to the questions addressed. I believe this can be a good example to encourage those who may also be facing competition in their research.
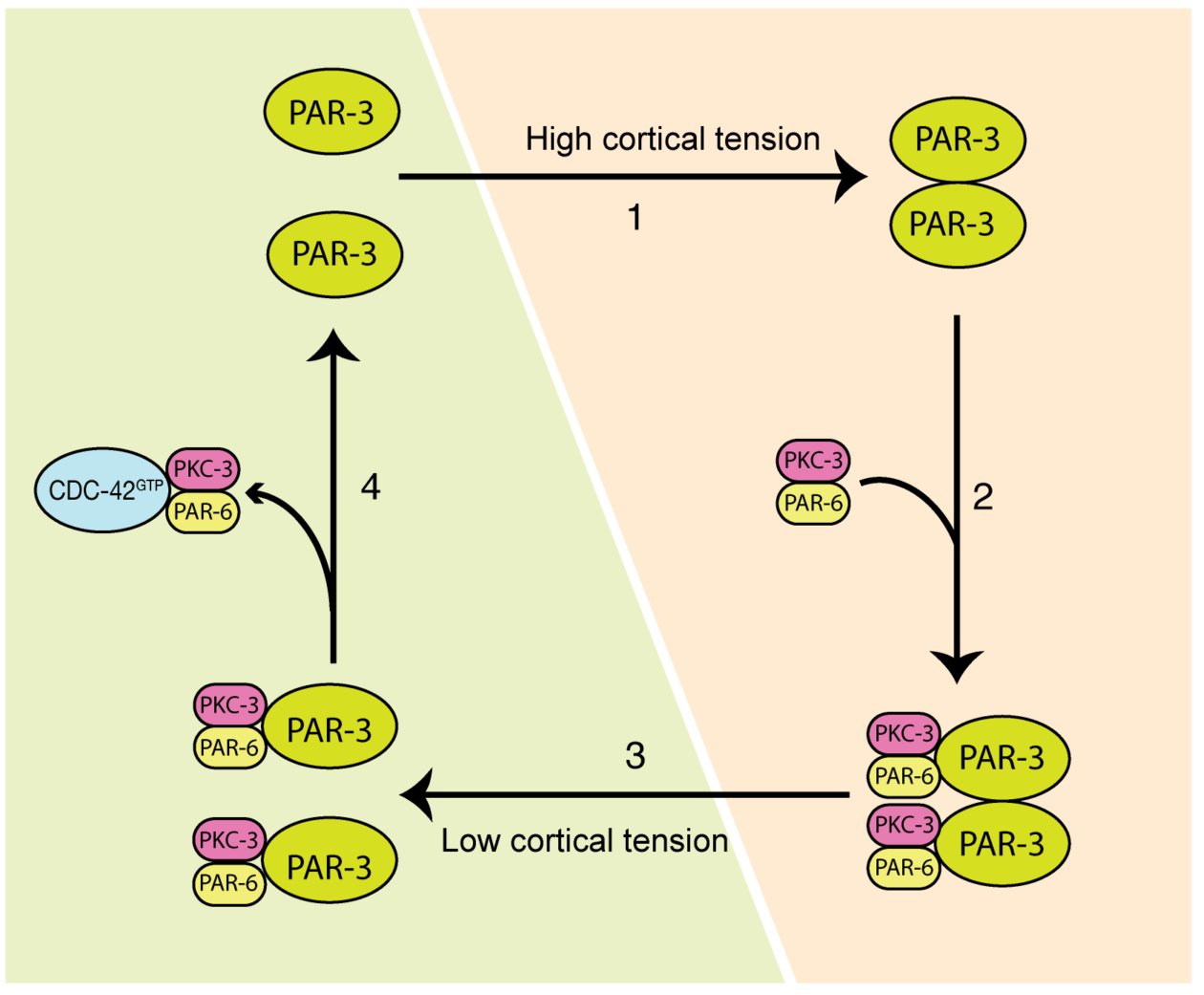
Several open questions about cell polarisation
The evolutionarily conserved PAR machinery is predominantly localized at the actomyosin cortex, which lies adjacent to the cell membrane. It is likely that the principle we uncovered is applicable to other types of tissue, and other organisms. For instance, it has been shown that self-assembly of the PAR-3 N’-terminal domain is critical for axon specification of neurons. In this case, the direct binding of the oligomerized domain to the microtubule promotes microtubule bundling and stabilization. It would be interesting to test if this formation of higher-order PAR-3 can be promoted by the activation of non-muscle myosin, which in turn changes local cortical tension. Another example is during Drosophila embryogenesis. Here, during the process of dorsal closure, actomyosin repeatedly assembles and disassembles in the epithelial amnioserosa cells. In this case, PAR-3 forms patches and overlaps with myosin at the apicomedial surface to promote actomyosin contraction. It is intriguing to explore whether myosin assembly induces structural changes in PAR-3, and whether this contributes to developmental morphogenesis. Finally, some follow-up questions remain elusive and need to be further addressed. What are the molecular mechanisms by which PAR-3 clustering or CDC-42 activity apply to compete for the PKC-3/PAR-6? How do aPARs clustering and actomyosin activity regulate and feedback to each other with a temporal-spatial precision during cell-cycle progression? It would be exciting to see emerging studies resolving more of the puzzles surrounding cell patterning in response to extrinsic and intrinsic stimulation in the near future.
References
Wang, S.C., Low, T.Y.F., Nishimura, Y., Gole, L., Yu, W. & Motegi, F. Cortical Forces and CDC-42 Control Clustering of PAR proteins for C. elegans Embyonic Polarization. Nat Cell Biol 19, 988–995 (2017).
Dickinson, D. J., Schwager, F., Pintard, L., Gotta, M. & Goldstein, B. A Single-Cell Biochemistry Approach Reveals PAR Complex Dynamics during Cell Polarization. Developmental Cell 42, 416–434.e11 (2017).
Rodriguez, J. et al. aPKC Cycles between Functionally Distinct PAR Protein Assemblies to Drive Cell Polarity. Developmental Cell 42, 1–35 (2017).
Chen, S. et al. Regulation of Microtubule Stability and Organization by Mammalian Par3 in Specifying Neuronal Polarity. Developmental Cell 24, 26–40 (2013).
David, D. J. V., Tishkina, A. & Harris, T. J. C. The PAR complex regulates pulsed actomyosin contractions during amnioserosa apical constriction in Drosophila. Development 137, 1645–1655 (2010).


 (7 votes)
(7 votes)
Congratulations Sam on the splendid findings and the publication . All the best for the future .
Was indeed an interesting read .
Thanks a lot bro. Looking forward to seeing your fabulous work being published soon!