Tills O, Spicer JI, Grimmer A, Marini S, Jie VW, Tully E, Rundle SD. 2018. A high-throughput and open-source platform for embryo phenomics. PLOS Biology, 16:1-19.
Background
A seasoned graduate student gave me some valuable advice at the outset of my PhD. She told me to FIND A BETTER WAY! We were both working on heterochrony – changes in the timing of developmental events. She had worked on between-species comparisons – I was about to start investigating within-species differences. The challenge in both research areas lies in its temporal nature – a poorly-timed lunch, or rest-room break and the developmental events of interest are literally a thing of the past. Development shows little regard for the diurnal rhythms of scientists either. Her advice drove me to develop a technology that would enable me to (occasionally) leave the lab. As a by-product, it produced a step change in the quality of the developmental data we were generating and, in turn, our understanding of development at the level of the whole-organism.
OpenVIM
OpenVIM is the result – a modular open-source video light microscope (Fig.1). Specifically tailored for automated time-lapse imaging of large numbers of aquatic embryos, for the duration of their development, or the experiment. The key with OpenVIM is that we can capture the dynamic process of embryonic development by generating realtime image sequences of each embryo. These acquisitions are repeated over the duration of the experiment (or their development). I started my career as a marine biologist, and despite my growing interest in development, my research continues to be set in the context of understanding how animals respond to changes in their environment. Therefore, environmental control was central and so this is also integrated and we can heat, cool and provide mixtures of gases during the course of an experiment.
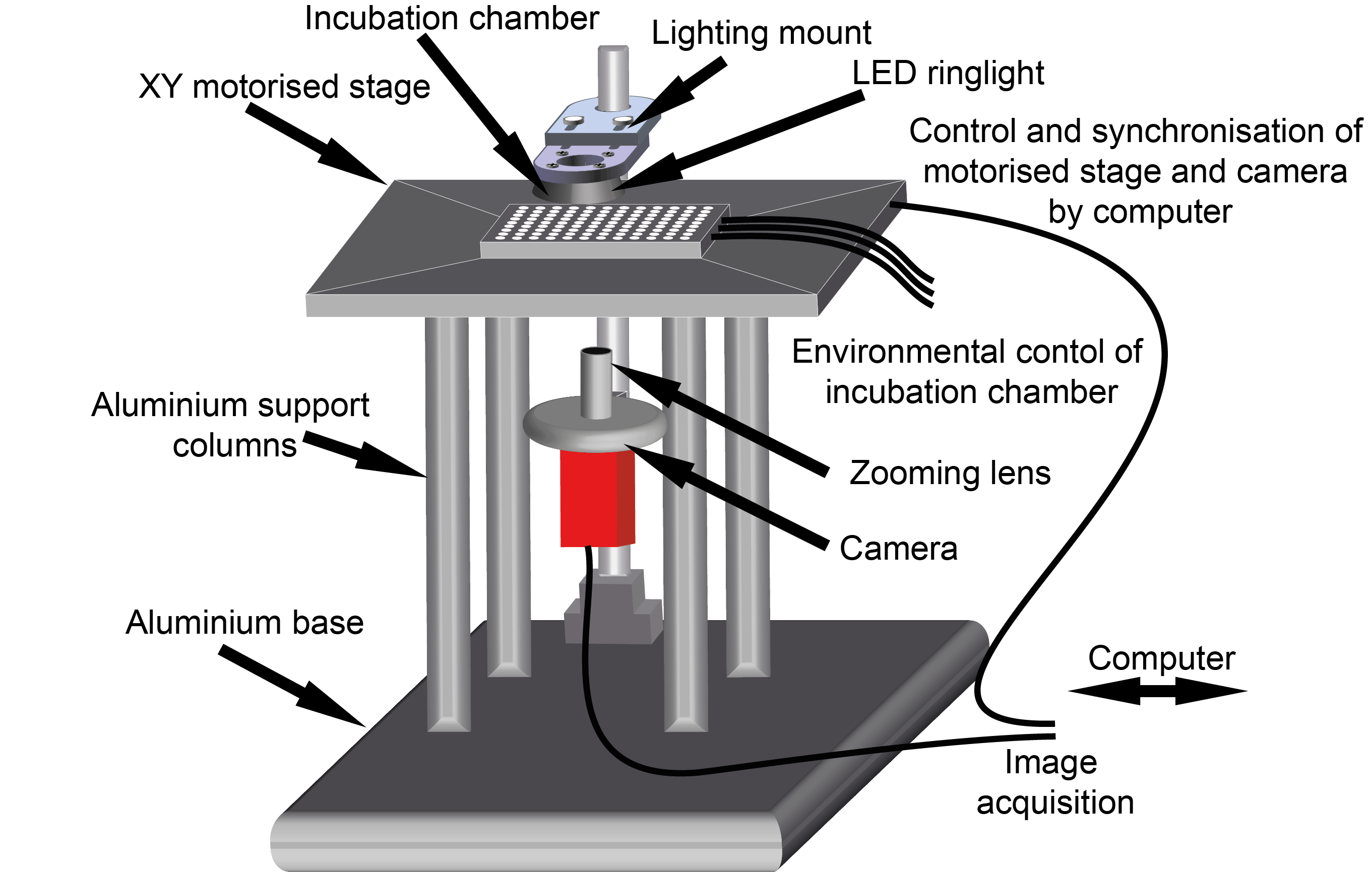
Fig.1. OpenVIM – Open-source video microscope, www.openvim.org.
OpenVIM produces many Terabytes (and hours) of video potentially leading to a serious bottleneck in its use. Fortunately a NERC Technology grant provided us the support to develop some complimentary analytical software that would relieve this bottleneck. We called this software EmbryoCV and it sits alongside OpenVIM and extracts as much biological information from these rich video time series as possible. The powerful combination of hardware and software we call EmbryoPhenomics – an open-source platform for phenomics in embryos.
EmbryoCV
EmbryoCV is a Python-based, high-throughput image analysis software. It sits alongside OpenVIM and automates the process of identifying embryos, measuring size, shape and movements all the way through to measuring physiological traits such as cardiac activity. This is performed on millions of frames, and hours of video, capturing both the real-time and developmental responses of embryos.
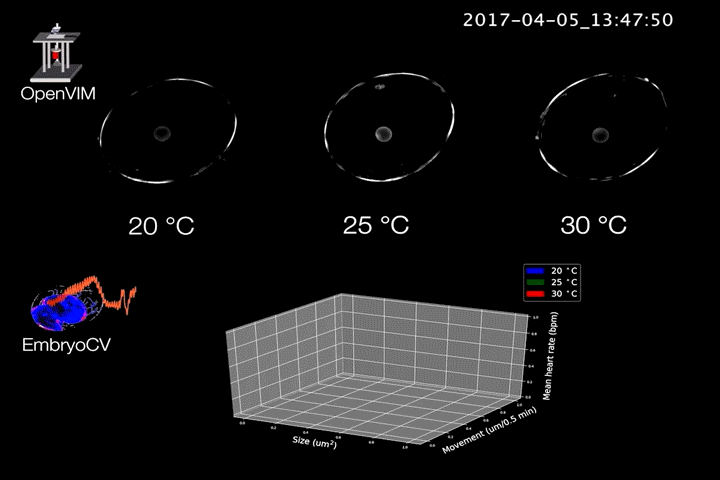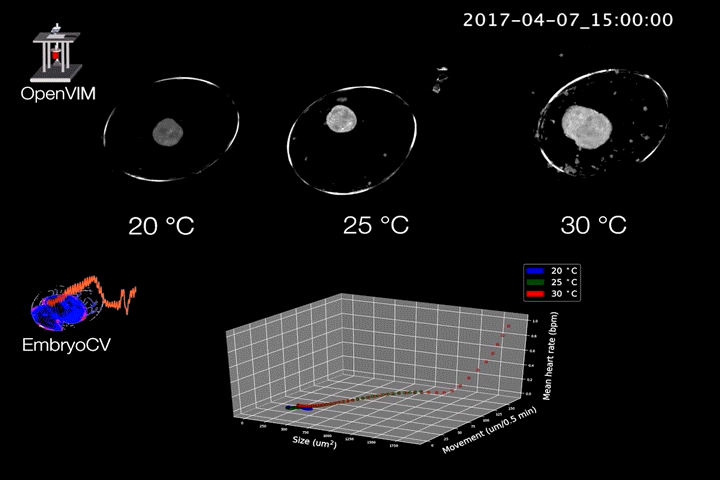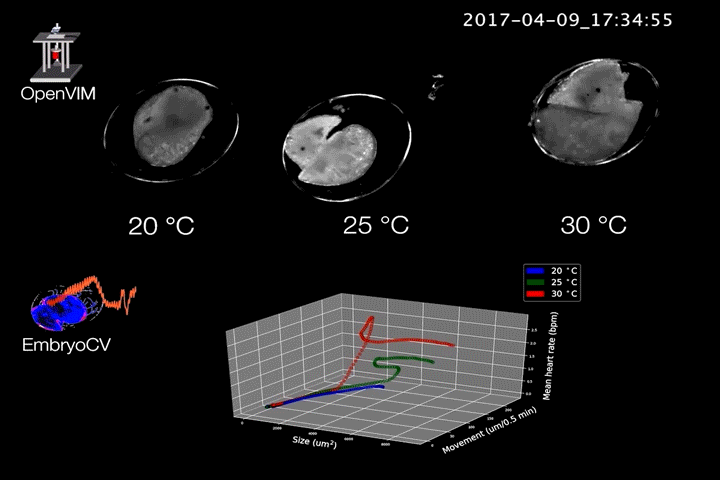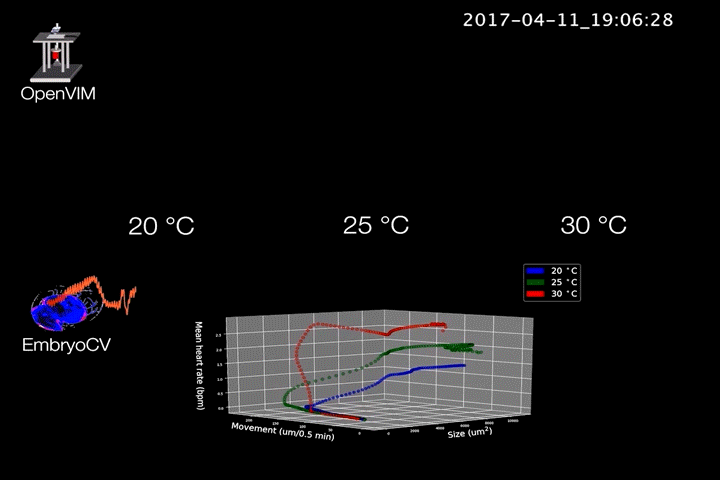
Fig.2. Video generated by OpenVIM of three Radix balthica embryos cultured at different temperatures. EmbryoCV generated measures of embryo size, movement and heart rate produced from 40 embryos at each temperature are shown in an XYZ plot.
A core component of EmbryoCV is the use of temporal fluctuations in pixel intensities in image sequences at different spatial resolutions. These signals can be used to make measurements of physiological traits such as heart rate. They can also be used to produce holistic measures such as lethal end points. to be identified.
EmbryoPhenomics
The combination of OpenVIM hardware and EmbryoCV software is called EmbryoPhenomics – an open-source platform for phenomics in embryos (Fig.2). Our recent paper describes the application of EmbryoPhenomics to perform four distinct climate-change themed experiments, using a freshwater gastropod and a marine amphipod – two species with very different types of development. In combination these experiments generated > 30 M images (approx 50 TB of image data). The EmbryoCV software was able to analyse > 93 % of these images.
Why phenomics?
David Houle
comments ‘
Organisms are fantastically complicated. Traditional biological intuition comes down to choosing an interesting character as the the object of study, from the essentially infinite number of characters that could be identified. Scientists who pick characters that are both biologically important and readily measured can obtain interesting results that resonate with others; those that do not pick so fortunately struggle‘.
Phenomics can be defined as the acquisition of high-dimensional phenotypic data on an organism-wide scale
Houle et al 2010. Technologies for high-throughput screening in phenomics address the challenge of data acquisition in much the same way that Next Generation Sequencing did in genomics – it reduces the limitations of traditional approaches, by applying high-throughput technologies to capture the complexity and interconnectedness of biological responses. Phenomics can provide a quality and quantity of data supportive of new approaches to understanding responses.
Biology is complex. Capturing the dynamic process of embryonic development using a handful of measures, perhaps chosen as much for their ease of measurement as their biological relevance is never going to be an optimal strategy. Nonetheless, this is the challenge faced by the majority of biologists, who do not possess the means to perform experiments to the scale that they would like.
Where next?
EmbryoPhenomics is open-source – both
OpenVIM hardware and
EmbryoCV software are freely accessible to people to adapt and modify to their own needs. The downstream data produced by EmbryoCV are vast and we are actively exploring different approaches to interrogate these, including deep learning. We are also streamlining the workflow of EmbryoPhenomics so that data are produced during, rather than after, an experiment.
Check out our
Vimeo Channel for some examples of the types of video and diversity of organisms that we work with.
More information on EmbryoPhenomics can be found in the paper or on these websites:
Dr Oliver Tills
Teaching and Research Associate, University of Plymouth
oliver.tills@plymouth.ac.uk
 (3 votes)
(3 votes)
 Loading...
Loading...



 (3 votes)
(3 votes)