BSDB Gurdon Studentship Report – Zhikai Li
Posted by Zhikai Li, on 9 December 2022
Soon after starting the first year of medical school, I had ambitions of undertaking substantial scientific research in future since biomedical research and clinical practice complement each other, but little idea about the nature of scientific research. On my search for a project, I came across the Gallop lab at the Gurdon Institute and was able to attend an interview with the lab where I presented a paper. I was then provisionally accepted as a summer student, and after a successful application for a BSDB Studentship, the project was given the go ahead.
The Gallop lab investigates actin signalling with emphasis on filopodia, regarding their macroscopic behaviour, molecular dynamics, and lipid composition. Actin biology had been mentioned in our first year courses but not explored further. I was therefore eager to research a relatively novel area for myself, and after discussion with Dr Gallop we decided on a project which would cater to both the labs and my interests.
The project would use embryos from the African clawed frog, Xenopus laevis, to study filopodia. The lab had recently discovered heterogeneity of actin regulatory molecules at filopodial tips, noting that filopodial extension and retraction rates in Drosophila myotubes forming myotendinous junctions follow a defined mathematical relationship, the Laplace distribution (Dobramysl et al. (2021)).
The distribution highlights how the poorly understood and apparently random biochemical processes in filopodia are still governed by some underlying order. The theoretical framework for our current understanding of filopodia dynamics, is that actin polymerisation is happens at the tips, with constant retrograde flow and actin depolymerisation occurring at the filopodium base (Mallavarapu and Mitchison, 1999). Therefore, a question my project sought to answer was whether other filopodia and actin incorporation at the tips also follow Laplace distributions in their dynamics. To answer this, I used leading edge mesendoderm (LEM) cells, isolated from stage 10.25 Xenopus gastrula and allowed to spread on a fibronectin matrix because of their accessibility and short incubation time. Additionally, the large cells at the 2-cell stage allow ease of injecting labelled membrane marker RNA (GAP-RFP) and labelled actin (with Atto-488). Any differences between the macroscopic behaviour of filopodia in cells in vivo compared ex vivo could provide insight into factors driving the filopodia behaviour.
If filopodia extension/retraction rates in Xenopus LEM cells fit a Laplace distribution, a further research question would be whether actin depolymerisation solely occurs at the filopodia base or if there may be actin depolymerisation at the filopodia tips too. Any findings can account for variations in the rates of filopodia extension and retraction.
TIRF (Total Internal Reflection Fluorescence) microscopy would be used to image filopodia. The membrane marker allows visualisation of the filopodia outline, and timelapses can then be analysed to track many filopodia over time using a plugin called ‘Filopodyan’ written in Fiji and R (Urbancic et al, 2017), producing graphs showing the distribution of filopodia extension and retraction rates. The labelled actin allows for photobleaching to answer the second research question, by tracking a stripe of photobleached actin.
The start of the project mostly involved learning techniques. I learned how to fertilise, treat, and inject embryos, take explants, and use the microscope. Because of the intricate nature of injecting embryos and taking explants, developing these skills were enjoyable but took time. I found explant-taking with eyebrow knives and hair-loops especially fun, reinforcing my desire to choose hands-on surgical specialties in future. Towards the end of the project, I was pleased to see the improvement in my aptitude for injecting embryos and taking explants.
The first experiment was in week 2, but it was found that the explants were not targeted enough, and a lot of vegetal endoderm persisted in the MaTek dishes used for imaging. There was also no actin fluorescence, so we hypothesised injecting GAP-RFP and labelled actin in separate needles, rather than a single, might allow actin to be introduced successfully. The second experiment was indeed more successful, as LEM cells with filopodia could be seen with some examples in Fig. 1 below.
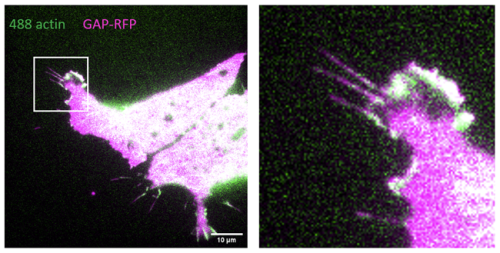
Fig. 1 – LEM cell with good filopodia, especially in the enlarged frame. Actin signal was weaker than membrane marker signal.
Further experiments focused on improving the actin signal intensity for more accurate filopodia segmentation in ImageJ. We tried switching wavelengths to use actin conjugated with a red dye (Alexa-568) and increasing the Atto-488 actin concentration, the latter of which proved more successful. Getting accurate and precise explants was also difficult, however refining the explant technique with help from Dr Gallop and my day-to-day supervisor, Julia Mason, improved the yield of LEM cells.
LEM cells had many mobile filopodia on the leading edge, however aside from segmentation problems due to low actin signal, overlapping filopodia were also inaccurately segmented by Filopodyan. A few cells from the final runs could be analysed fully in ImageJ, generating tables which were fed into the Filopodyan script in R and producing graphs that approximated a Laplace distribution of filopodia extension and retraction rates.
Finally, to address the project specifically, the script had to be rewritten to plot a logarithmic scatter plot instead of a histogram. The best fit lines still need integrating into the script, and this along with perfecting the experimental procedure could form the basis for another short project to finish my work on the Laplace distribution and answer the second part of the research question on actin dynamics.
Overall the project has been invaluable, not only in developing hard skills like working with basic bench equipment, manipulating embryos and imaging, but also in highlighting the unpredictable nature of scientific research. Completing this project has better prepared me for future research opportunities, which I will be seeking, as well as allowed me to meet excellent colleagues which have been of great help, for which I express my deepest gratitude.
References:
Dobramysl U, Jarsch IK, Inoue Y, Shimo H, Richier B, Gadsby JR, Mason J, Szałapak A, Ioannou PS, Correia GP, Walrant A, Butler R, Hannezo E, Simons BD, Gallop JL. Stochastic combinations of actin regulatory proteins are sufficient to drive filopodia formation. J Cell Biol. 2021 Apr 5;220(4):e202003052.
Mallavarapu A, Mitchison T. Regulated actin cytoskeleton assembly at filopodium tips controls their extension and retraction. J Cell Biol. 1999 Sep 6;146(5):1097-106.
Urbančič V, Butler R, Richier B, Peter M, Mason J, Livesey FJ, Holt CE, Gallop JL. Filopodyan: An open-source pipeline for the analysis of filopodia. J Cell Biol. 2017 Oct 2;216(10):3405-3422.


 (7 votes)
(7 votes)