Behind the paper story: ‘How heterotypic cis/trans factors drive stomatal cell fate commitment’
Posted by EUN-DEOK KIM, on 16 March 2023
Dr. Eun-Deok Kim and Professor Keiko Torii from The University of Texas at Austin, USA, have recently published an article in Nature Plants. The article discusses how heterotypic cis/trans factors drive cell fate commitment during stomatal development. The Node asked us to provide a behind the scenes look at how the story came together.
Nature Plants: Dynamic chromatin accessibly deploys heterotypic cis/trans acting factors driving stomatal cell fate commitment.
How did you get started on this project?
One of the fundamental questions in the development of multicellular organisms is how cells with the same genomic content acquire distinct identities. Recent research has highlighted the importance of chromatin in regulating gene expression 1-3. Nevertheless, the precise mechanisms by which combinations of cis-regulatory elements (CREs) and trans-acting factors interact to promote cell-fate determination remains unclear. Likewise, we still don’t fully understand the relationship between chromatin architecture and transcription factor binding, leading to cell fate specification during development remains unclear. To address this question, we are using plant stomatal differentiation as a model system.
Can you summarise your findings?
This study is a big step forward in understanding how stomatal cell lineages progress and differentiate, thanks to the use of a multi-omics approach, which includes epigenomics, genetics, and biochemistry. By uncovering the unique combinatorial cis/trans-regulatory codes that drive stomatal cell lineage progression, we’ve made a breakthrough in this field. The early stomatal precursor state is initiated by SPCH and terminated by MUTE, two sister bHLH proteins 4,5. However, how these proteins switch the cell state from proliferation to differentiation has remained a mystery until now. Using specialized techniques such as stomatal-lineage specific ATAC-seq (INTACT-ATACseq), ChIP-seq, and unbiased transcription factor screens, we identified bHLH (E-box) and BBR/BPC (GAGA-repeat) motifs as unique Co-CREs that signify the early stomatal precursor state 6.
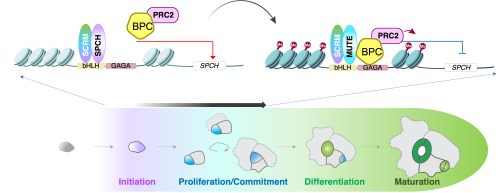
Figure 1. adapted from Kim et al. 2022, which is licensed under a Creative Commons Attribution 4.0 International License
Additionally, we uncovered the exact mechanism by which MUTE drives stomatal cell-fate commitment. MUTE (with its partner bHLH, SCREAM 4,5,7,8) binds to target DNA (E-box) and can act as a transcriptional activator. At the same time, MUTE directly associates with BPC proteins (binding to the nearby BBR/BPC GAGA repeat DNA motif of the Co-CREs), which recruit Polycomb Repressive Complex2 (PRC2) 9,10and repress the SPCH locus via deposition of repressive histone marks. Depending on the cell stage, specific heterotypic transcription factor interactions drive the switch from proliferation to the commitment stage. We have shown through genetic, biochemical, biophysical, and in vivo functional evidence that the dysregulation of this mechanism leads to hyper-proliferation or lack of stomatal cell lineages. By integrating chromatin landscape dynamics with molecular mechanistic details, our work has shed light on the novel role of disparate pairs of cis- and trans-acting factors in shaping cell-fate commitment during specialized cell-type differentiation.
Do you think that versatile heterotypic transcription factor interactions are likely to play a key role in regulating differentiation in other systems?
A diverse set of heterotypic TF complexes has been observed during cardiogenesis, hematopoiesis, and myogenesis (the development of muscle tissue)11-16 For instance, certain TFs like T-box TF TBX5, the homeodomain TF NKX2-5 and the zinc finger TF GATA4 form heterotypic interactions that coordinate cardiac gene expression, differentiation, and morphogenesis. These interactions also limit the potential of these TFs to bind to irrelevant regulatory elements in a given context. Myo-D, a myogenic basic helix-loop-helix (bHLH) protein, and the myocyte enhancer MADS domain TF MEF2 can cooperatively regulate the initiation of myogenesis as another example of a heterotypic complex. I strongly believe that heterotypic interactions not only between transcription factors but also epigenetic regulators play a crucial role in regulating differentiation in many other biological systems.
When doing the research, did you have any particular result or eureka moment that has stuck with you?
Yes, upon observing the first inducible BPC phenotype and the interaction between MUTE-BPC but not SPCH-BPC. I collaborated with Bridget Fitzgerald, then a research technician, and Hyemin Seo, a graduate student in Keiko Torii lab, and we were all very excited to discover that the inducible overexpression phenotype of BPC suggested the repression of SPCH and a cell-state-specific interaction.
And what about the flipside: any moments of frustration or despair?
During the pandemic, like many others, I didn’t have enough time to work on this project. Additionally, generating multiple transgenics and crosses was time-consuming and demanding.
Where will this story take the lab?
Our next goal is to investigate how the combination of cis- and trans-acting factors, as well as chromatin dynamics, mutually impact gene expression to promote cell fate specification at the single-cell resolution. Furthermore, our study indicates that the timely upregulation of MUTE could play a critical role in driving the switch of the epigenomic landscape towards stomatal differentiation. Previous molecular-genetic research has suggested that the HD-ZIP IV family and other transcription factors may promote MUTE expression, but the direct mechanism of action has not been explored. Nevertheless, the future question to address is how heterotypic TF groups, complexed with epigenetic modifiers, differentially guide the developmental progression at the atomic level.
What next for you after this paper?
I am currently conducting research on the progression of stomatal lineage cells and the active role of chromatin dynamics in determining stem cell fate specification, differentiation, and maintenance. I am now seeking tenure-track positions to establish my own research group and continue to explore these topics further.
References
- Cusanovich, D. A. et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 555, 538-542, doi:10.1038/nature25981 (2018).
- Cusanovich, D. A. et al. A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell 174, 1309-1324.e1318, doi:10.1016/j.cell.2018.06.052 (2018).
- Marand, A. P., Chen, Z., Gallavotti, A. & Schmitz, R. J. A cis-regulatory atlas in maize at single-cell resolution. Cell 184, 3041-3055.e3021, doi:10.1016/j.cell.2021.04.014 (2021).
- MacAlister, C. A., Ohashi-Ito, K. & Bergmann, D. C. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537-540, doi:10.1038/nature05491 (2007).
- Pillitteri, L. J., Sloan, D. B., Bogenschutz, N. L. & Torii, K. U. Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501-505, doi:10.1038/nature05467 (2007).
- Kim, E.-D. et al. Dynamic chromatin accessibility deploys heterotypic cis/trans-acting factors driving stomatal cell-fate commitment. Nature Plants, doi:10.1038/s41477-022-01304-w (2022).
- Kanaoka, M. M. et al. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell 20, 1775-1785, doi:10.1105/tpc.108.060848 (2008).
- Ohashi-Ito, K. & Bergmann, D. C. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18, 2493-2505, doi:10.1105/tpc.106.046136 (2006).
- Xiao, J. et al. Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat Genet 49, 1546-1552, doi:10.1038/ng.3937 (2017).
- Wu, J. et al. Spatiotemporal Restriction of FUSCA3 Expression by Class I BPCs Promotes Ovule Development and Coordinates Embryo and Endosperm Growth. Plant Cell 32, 1886-1904, doi:10.1105/tpc.19.00764 (2020).
- Cao, J. et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361, 1380-1385, doi:10.1126/science.aau0730 (2018).
- Luna-Zurita, L. et al. Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis. Cell 164, 999-1014, doi:10.1016/j.cell.2016.01.004 (2016).
- Ogata, K., Sato, K. & Tahirov, T. H. Eukaryotic transcriptional regulatory complexes: cooperativity from near and afar. Curr Opin Struct Biol 13, 40-48, doi:10.1016/s0959-440x(03)00012-5 (2003).
- Glasmacher, E. et al. A Genomic Regulatory Element That Directs Assembly and Function of Immune-Specific AP-1-IRF Complexes. Science 338, 975-980, doi:10.1126/science.1228309 (2012).
- Rothenberg, E. V. Encounters across networks: Windows into principles of genomic regulation. Mar Genom 44, 3-12, doi:10.1016/j.margen.2019.01.003 (2019).
- Rothenberg, E. V. Logic and lineage impacts on functional transcription factor deployment for T-cell fate commitment. Biophys J 120, 4162-4181, doi:10.1016/j.bpj.2021.04.002 (2021).


 (2 votes)
(2 votes)