An After Thought to Evolution: Exceptional ways of Controlling Gene “Expression”
Posted by Linda, on 3 May 2011
More and more, the central dogma is becoming well, dogged, for being a dogma at all. As humans, we have 3 billion nucleotides. Only 1% of it makes up our protein coding genes, which led to the development of the central dogma: DNA is transcribed to RNA and translated into proteins. During undergrad, we’re taught that transcription factor proteins bind to sequences in the DNA to either enhance or shut off transcription, thereby discontinuing protein production. Gene regulation seems to follow certain pathways and rules itself.
Then, 20 years ago RNAi was discovered, a whole new field where RNA can also regulate protein production. Generally, small RNAs repress protein coding genes by either causing their mRNAs to decay or inhibiting their translation into protein. They can be naturally produced for the sake of controlling the native genes, following a similar central dogmatic pathway: DNA is transcribed to a precursor RNA, which is converted into teeny RNA strands. These strands are selected by Argonaute proteins (AGOs) and used to target specific protein coding genes to halt their expression.
The progression from DNA to RNA to “something”, is like the foundation of genetics. And gene regulation is about how to control that progression. Evolution develops these elegant pathways to produce “something”, and then it evolved ways to control the pathway.
Well, not always.
 Sometimes you can get “unintelligent design.”
Sometimes you can get “unintelligent design.”
Which is what a student joked, at a journal club meeting I’d attended today. The meeting started off as a presentation of a recently published article that assigned a role to an AGO protein in Arabidopsis. It ended up as a digression on evolution and how we perceive it, among other things.
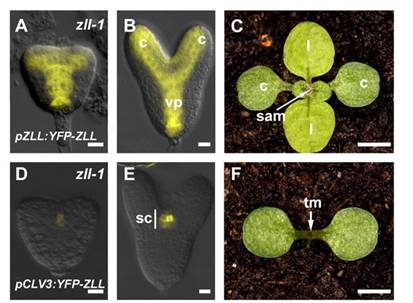
The protein of interest was AGO10, which is somehow involved in miRNA/small RNA directed repression of target genes. But how it exactly it does this, was hazy to us, the observers. By inference, previous studies on its loss of function mutant, ago10/zwille/pinhead had deformed shoot apical meristems. After embryogenesis, the meristems at the junctions of leaves are where new ones form. In the mutants, the meristem is the size of pinhead, smaller than normal (Pictured in F, on the left, versus wild type in C, by Tucker et al., 2008). The protein expression of some transcription factors & signalling peptides changed in the ago10 mutants as well. But what does AGO10 specifically do?
(Picture: A, B = AGO10 expression in the yellow YFP areas of the embryo, at heart stage & torpedo stage. CLV, a stem-cell specific signalling peptide. its expression is weak is in the embryo ago10, as pictured in D, E. (it’s not strong in WT either though) CLV needs AGO10 to be maintained past embryogenesis, in the meristem Tucker et al. 2008).
One group discovered that AGO10 doesn’t really help in repression. It actually indirectly causes the activation of target genes. AGO10 is a decoy for the main AGO1, that performs 90% of miRNA directed repression in Arabidopsis plants. However, it’s a decoy for one miRNA alone (so it seems). It binds and sequesters miR166/165, a well studied miRNA that is deeply conserved in mosses and higher plants. By sequestering it, miR166/65 is prevented from binding to AGO1 and regulating its targets (HD ZIP proteins). Effectively it’s as if miR166/65, the silencer, has been silenced itself. By a massive protein no less.
Oddly, AGO10 is most abundant in one tissue type, the developing embryo. So it only catches up all the miR166/65 population in that one tissue. If it doesn’t, as suggested by ago10, then the embryo develops into mutant plant. 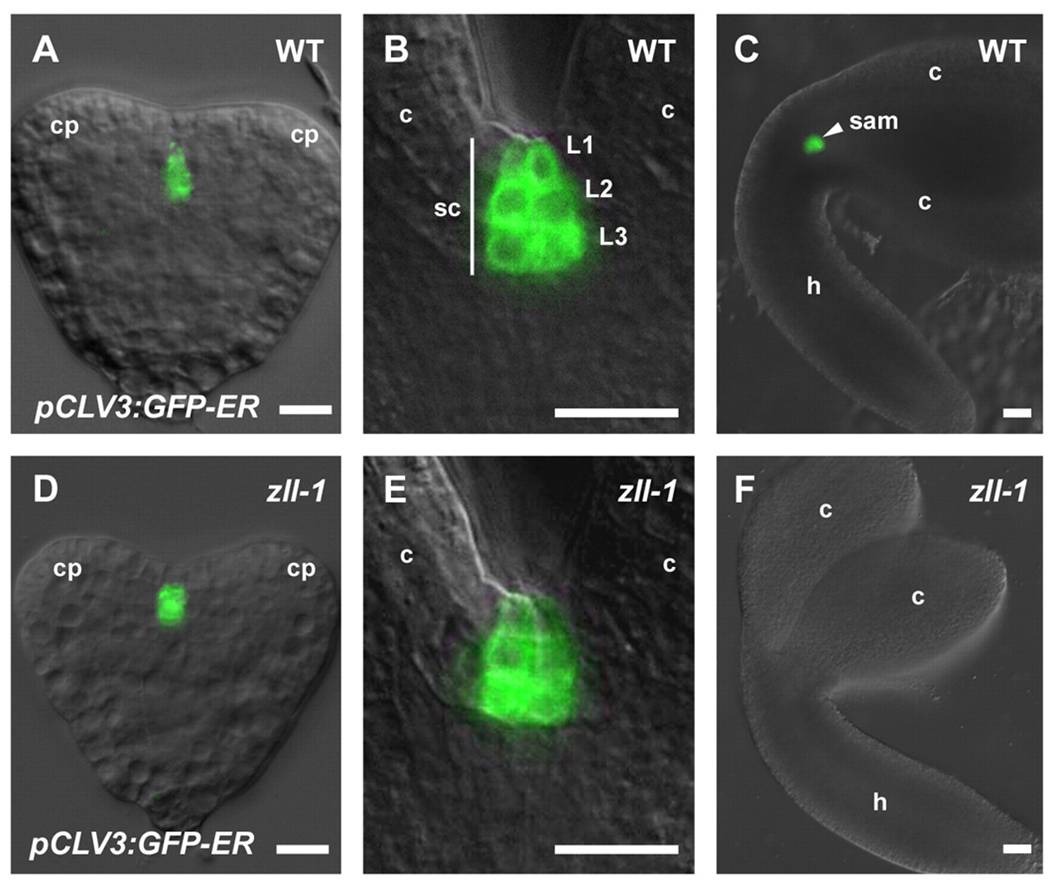
But why would plants bother developing this? Couldn’t nature have just evolved so that miR166/65 isn’t transcribed in the embryo? So that some transcription factor or miRNA shuts it off. Like, why produce something and then evolve a whole protein to muffle its activity? It’s like..evolution forgot to turn off the tap or shut the door, and created a massive sink or club bouncer instead. Energy wise, it seems a waste though. Evidently there probably is an unknown purpose.
The authors of the paper suggest that miR166 is required in specific cells, but not all cells of the developing embryo. So miR166 is allowed to be expressed but AGO10 prevents it from being spread elsewhere..assuming that miRNAs can travel…
(A-C: WT, CLV expression is maintained throughout embryogenesis and meristem development. Without AGO10 in D-F, its expression disappears).
![]() It’s funny. We have these commonly accepted notions about gene regulation engrained in us from years of training. With the result that when we’re faced with something altogether new, we’re not sure what to make of it. We’re not even sure if we can accept it, but there it is. It’s like realizing black swans with ghoulish red eyes exist in Australia.
It’s funny. We have these commonly accepted notions about gene regulation engrained in us from years of training. With the result that when we’re faced with something altogether new, we’re not sure what to make of it. We’re not even sure if we can accept it, but there it is. It’s like realizing black swans with ghoulish red eyes exist in Australia.
Sources/References:
Lab discussion with students at the Research School of Biology at ANU, led by PIs Tony Millar and Iain Searle.
Zhu H, Hu F, Wang R, Zhou X, Sze SH, Liou LW, Barefoot A, Dickman M, & Zhang X (2011). Arabidopsis Argonaute10 Specifically Sequesters miR166/165 to Regulate Shoot Apical Meristem Development. Cell, 145 (2), 242-56 PMID: 21496644
Pictures:
Tucker MR, Hinze A, Tucker EJ, Takada S, Jürgens G, & Laux T (2008). Vascular signalling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development (Cambridge, England), 135 (17), 2839-43 PMID: 18653559
 Flickr CC by Kyknoord
Flickr CC by Kyknoord


 (5 votes)
(5 votes)