Behind the paper: A transcriptomic hourglass in brown algae
Posted by Jaruwatana Sodai Lotharukpong, on 12 November 2024
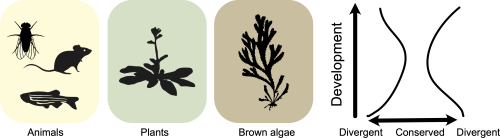
Brown algae are a group of complex multicellular eukaryotes, unrelated to animals, plants and fungi. It follows that brown algae evolved the process of multicellular development independently, offering a unique opportunity to investigate shared principles underlying developmental evolution across the tree of life. One such principle is the hourglass model of embryo evolution. The hourglass model describes a pattern of evolutionary conservation and divergence during embryogenesis, where the divergent earlier and later stages are bridged by a conserved mid-embryonic period (Duboule 1994). Such hourglass patterns have been observed in animal, plant and fungal development (Drost et al. 2017). But what about brown algae?
In our recent paper, “A transcriptomic hourglass in brown algae” (Lotharukpong et al. 2024), we asked a simple question: does brown algal development follow the hourglass model? By profiling the developmental transcriptome in two Fucus species, we observe an hourglass pattern in brown algal embryogenesis (Fig 1). The conserved mid-embryonic period is underpinned by the reduced expression of evolutionarily younger genes and the presence of more broadly expressed, potentially pleiotropic genes. We also explored the transcriptome across life cycle stages in brown algae with differing levels of morphological complexity. Crucially, in morphologically simple Ectocarpus species without canonical embryos, multicellular development itself appeared to constrain transcriptome evolution, suggesting how such embryo hourglass patterns may have emerged in the first place (at least in brown algae). Overall, this work gives evidence for the hourglass model as a general principle underlying developmental evolution across the tree of life.
But as we all know; a lot went on behind the paper.
Sampling and growing brown algal species:
To test the hourglass model in brown algae, we first had to literally find morphologically complex brown algae that undergo embryogenesis (Fig 2). Between France and Germany, Rémy Luthringer (co-author) had to go collect reproductively mature adults to produce embryos and cultivate them in the lab. It was a challenge to culture Fucus embryos beyond the earliest embryonic stages, since they require a lot of care to ensure healthy growth. This was all the more difficult for species such as F. distichus, which required up to half a year to reach the latest embryonic stage used in the study.

Bioinformatics in the spotlight:
We then faced several hurdles on the bioinformatics side when piecing this study together, which required a sustained push on software development. In particular, we wanted to infer the evolutionary age of each gene, which is needed for computing the transcriptome age index (a key metric for evolutionary transcriptomics). Existing approaches were not suitable nor computationally scalable to current large databases such as the National Center for Biotechnology Information (NCBI) non-redundant (nr) database and did not account for potential database contaminations among other confounding factors in gene age inference. We therefore teamed up with Josué Barrera-Redondo to create GenEra (Barrera-Redondo et al. 2023), a wrapper around the fast and sensitive pairwise sequence aligner DIAMOND v2 (Buchfink et al. 2021), which brought down the search time from months to days. This finally allowed us to infer the gene age in a timely manner for any eukaryotic genome, including the species we used in the hourglass study (Fucus serratus, Fucus distichus, Ectocarpus sp., Laminaria digitata and Saccorhiza polyschides).
On top of this, we extended the functionalities of R packages for evolutionary transcriptomics, such as myTAI (Drost et al. 2018), to accommodate our analyses. New and existing functions in myTAI enabled us to distinguish evolutionary signals from random noise. We hope these efforts will be useful for the wider community interested in asking evo-devo questions using transcriptomic data.
What’s next for the story?
It is an exciting time for brown algal research. A recent study has provided several dozen genome assemblies across major groups of brown algae (Denoeud et al. 2024). Furthermore, functional genetics is now a possibility for multiple brown algal species, fuelling recent findings such the independent evolution of HMG domain genes as a male sex-determining factor (Luthringer et al. 2024). There is also a drive to generate transcriptomic data to understand cell-type and tissue evolution in brown algae, which also evolved cell-types and tissues independently from other complex multicellular eukaryotes. There is still much more to come!
References
Barrera-Redondo J, Lotharukpong JS, Drost H-G, Coelho SM. 2023. Uncovering gene-family founder events during major evolutionary transitions in animals, plants and fungi using GenEra. Genome Biol. 24:54.
Buchfink B, Reuter K, Drost H-G. 2021. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18:366–368.
Denoeud F, Godfroy O, Cruaud C, Heesch S, Nehr Z, Tadrent N, Couloux A, Brillet-Guéguen L, Delage L, Mckeown D, et al. 2024. Evolutionary genomics of the emergence of brown algae as key components of coastal ecosystems. :2024.02.19.579948. Available from: https://www.biorxiv.org/content/10.1101/2024.02.19.579948v2
Drost H-G, Gabel A, Liu J, Quint M, Grosse I. 2018. myTAI: evolutionary transcriptomics with R. Bioinformatics 34:1589–1590.
Drost H-G, Janitza P, Grosse I, Quint M. 2017. Cross-kingdom comparison of the developmental hourglass. Curr. Opin. Genet. Dev. 45:69–75.
Duboule D. 1994. Temporal colinearity and the phylotypic progression: a basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Development 1994:135–142.
Lotharukpong JS, Zheng M, Luthringer R, Liesner D, Drost H-G, Coelho SM. 2024. A transcriptomic hourglass in brown algae. Nature 635:129–135.
Luthringer R, Raphalen M, Guerra C, Colin S, Martinho C, Zheng M, Hoshino M, Badis Y, Lipinska AP, Haas FB, et al. 2024. Repeated co-option of HMG-box genes for sex determination in brown algae and animals. Science 383:eadk5466.


 (3 votes)
(3 votes)