BSDB Gurdon Studentship Report – Ewa Ozga
Posted by Ewa Ozga, on 19 December 2022
Exploring the role of planar cell polarity in the regulation of the patterning of human axial progenitors
I first became interested in stem cell biology and development while studying Medical Sciences at the University of Edinburgh. During the lockdown, I attended a very inspiring seminar in which Dr Guillaume Blin discussed in-vitro models of developmental patterning. Despite being unable to join the lab in my second year, I explored the topic, and we discussed some ideas from the literature. The BSDB summer studentship provided me with a perfect opportunity to explore these ideas in a lab setting. This summer, I got a chance to work alongside an exceptional team in the Blin lab on my own project at the Centre for Regenerative Medicine (CRM).
I have been interested in the role of planar cell polarity (PCP) in health and disease and decided to investigate its role in developmental patterning. PCP dysregulation can play a significant role in cancers and congenital disorders (Wang, de Marco, Capra and Kibar, 2019). In particular, in congenital malformations of spinal structures, such as neural tube defects (NTDs), where aetiology is still to be elucidated (Chen et al., 2018). A proportion of individuals with NTDs (Chen et al., 2018) and idiopathic scoliosis (Wise et al., 2020) have mutations in some of the core components of the PCP pathway, namely Wnt11, Vangl1 and Vangl2, and Celsr. These components localise asymmetrically within the cell to define cell polarity along the epithelial plane (Butler and Wallingford, 2017).
This pathway is involved in early development, notably when spinal progenitors are established in a structure called the anterior primitive streak (APS) (Andre et al., 2015). As these transient and rapid events occur within a complex 3D environment in-utero, much remains to be understood about cell fate decisions in the APS (Wymeersch et al., 2021). One way to study these processes in a human context is to use human embryonic stem cells to model early development (Blin, 2021). During my project, I worked on a novel in-vitro model that mimics the early stages of human axis elongation. I used this system to test the hypothesis that PCP regulates the patterning and balanced proportion of early spinal progenitors in humans.
My experimental strategy consisted in confining hESC onto custom-made micropatterns and treating the colonies with a spinal fate-inducing medium. In these conditions, hESC self-organise into spatial domains of cell fates and initiate axial growth. I first tested a set of antibodies directed against PCP components. These antibodies did not provide a specific signal, but I obtained images that looked magical, so I was not too disappointed! Next, I decided to use small molecule inhibitors to perturb the PCP pathway. I inhibited either cytoskeleton remodelling downstream of PCP or the secretion of PCP ligands. When I stained for neurectoderm and endodermal cell fate markers, I observed a very severe patterning defect when cytoskeleton remodelling was inhibited (Figure 1).
I was very excited with these clear preliminary results and I wish I had the time to perform additional experiments that would demonstrate the involvement of PCP more specifically, such as PCP components staining and knockdown experiments. I have also assisted my colleagues in testing new micropatterning methods and creating cell lines using transfection methods.
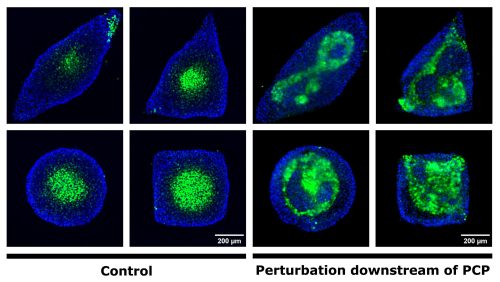
Figure 1. Perturbation of cytoskeletal remodelling downstream of PCP leads to neurectoderm patterning defects. Immunostaining of micropatterned colonies 48h after the induction of differentiation. The endoderm is shown in blue. Neurectodermal cells (green) cluster at the centre in the control condition (a-d) regardless of colony shape, while patterning is perturbed in the treated condition (e-h).
Thinking back to my first days in the lab I was very excited to work alongside experienced scientists but also felt anxious as I needed to use techniques that I was not yet familiar with. During my first week, I worked out how to employ the techniques routinely used in the lab and presented my initial research plan to discuss my approach. Once I became acquainted with the protocols, they became a natural part of my routine. For example, I became proficient in micropatterning, a microfabrication method that makes it possible to standardise the size and geometry of hESC colonies (Blin, 2021). This project also allowed me to try several imaging techniques and perform image analysis using Nessys (Blin, 2019) and PickCells. I really enjoyed organising, sharing, and optimising the protocols in an online lab book. I am so happy I could contribute and that my designs are now used by the lab. We have also collaborated with a computational lab and worked on shared scripts in python.
During my internship, we also attended the Mammalian Synthetic Biology congress taking place in Edinburgh. It was a great opportunity to hear about novel data and techniques from researchers around the world and discuss how we could apply them in our lab. Networking and making friends in a professional environment like the CRM gave me the opportunity to gain unique perspectives from postgraduate students and researchers from various groups. Presenting and discussing my ideas and data allowed me to gain more insight into the dynamics of working in research and academia. These 8 weeks inspired and prepared me to pursue a career in science.
My greatest gratitude goes to my amazing supervisor, Guillaume, for everything that he has done for me. I am so grateful to Miguel for always being there for me, Heather for caring for all of us, and Fatma for the priceless moral support (all below).

Figure 2. The Blin lab (from left): Me, Fatma, Miguel, Guillaume, and Heather.
I want to express my gratitude to the amazing people at the CRM who were always keen to help and explain their experiments with so much passion. Many thanks to the BSDB for giving students like me such a wonderful opportunity to kickstart research careers. I would recommend all students to start looking for a lab they would be interested in doing a BSDB-founded internship in!
References


 (No Ratings Yet)
(No Ratings Yet)