BSDB Gurdon Summer Studentship Report (1)
Posted by BSDB, on 27 November 2014
In 2014, the BSDB has initiated the Gurdon Summer Studentship program with the intention to provide highly motivated students with exceptional qualities and a strong interest in Developmental Biology an opportunity to engage in practical research. The 10 successful applicants spent 8 weeks in the research laboratories of their choices, and the feedback we received was outstanding. Please, read the student report kindly sent to us by Benedetta Carbone.
I am Benedetta, an undergraduate student studying Molecular Genetics at the University of Edinburgh. This summer I had the amazing opportunity to spend 8 weeks in a research lab working on stem cells.
It all started out because after 3 years of studying Biology and an interest in building a career in research, I didn’t really know what the job of a full time researcher looked like. So, encouraged by University professors, I started investigating fields and topics that I had liked the most during my studies.
I was doing some research on induced pluripotent stem cells when I came across Dr. Kaji’s lab at the Centre for Regenerative Medicine in Edinburgh. Their work focuses on the Biology of Reprogramming, the molecular changes that occur in reprogramming cells and how the process can be further understood and improved.
I contacted him to ask if I could join the lab for the summer. He offered me a place for an internship and designed a project focusing on DNA adenine methyltransferase identification (DamID). DamID is a technique based on the bacterial protein DNA adenine methyltransferase (Dam). This protein recognises GATC sequences and methylates position 6 of the adenine. DamID works by tethering Dam to a DNA-binding protein. The target protein binds to DNA localising Dam to the same sites within the genome. Thus, Dam has a higher chance of methylating GATC sequences around the binding site of the target protein.DamID is hence used to detect DNA-binding sites for target proteins, producing data similar to ChIP-seq. As a result, DamID does not require the use of any antibody, reducing the amount of time necessary to process DNA and the amount of cells required as starting material.
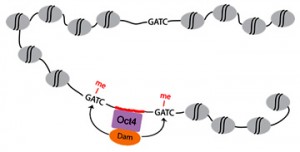
One of the PhD students in Dr. Kaji’s Lab, Luca Tosti, has been working with DamID and the aim of my summer project was to test the DamID technology by generating my own Dam expressing cell line to investigate Oct4 binding sites in mouse Embryonic Stem Cells (mESCs).
And so my internship began. At first I was a bit overwhelmed and I found myself confused by very simple things. It quickly got a lot better, thanks to everyone in the lab (especially Luca!): they were all very friendly, helpful and patient!
I started by cloning the expression vectors and I gained experience in basic molecular biology techniques such as restriction enzyme digestions and ligations, bacterial transformations, extraction of plasmid DNA and gel electrophoresis. I also familiarised myself with the ApE software to design cloning strategies.
I generated several vectors to test different levels of Dam expression in order to get a good signal-to-noise ratio. Indeed, one of the main issues related to this technology is the high background signal arising from the intrinsic DNA-binding activity of the Dam protein.
Afterward, I was introduced to tissue culture where I worked with mouse ESCs (mESCs). I learned how to maintain mESCs in culture and how to passage them when confluent. I also gained experience on plasmid transfection, picking colonies, freezing and thawing cells. I transfected mESCs with the vectors I had previously generated and I used the antibiotic resistance cassette I had cloned into the constructs to select for cells expressing the transgene.
As a part of the project, I also wanted to check how the adenine methylation signal changes when mESCs differentiate, and for this purpose I performed retinoic acid (RA) differentiation of mESCs. After removing Leukaemia Inhibitory Factor (LIF, a signalling molecule important for maintaining the undifferentiated state of mESCs) and adding RA to the medium (for 9 days), I was able to observe significant morphological changes.
I then went back to the lab bench: I learned how to extract RNA and how to perform RT-qPCR to determine gene expression patterns. Using this approach I could confirm that differentiated cells had switched off pluripotency markers. I also learned how to extract genomic DNA and how to use enzymatic digestion coupled to qPCR to quantify the methylation levels of DNA around GATC sequences. My results were very encouraging. I managed to get good expression of the Dam-Oct4 fusion protein and to observe good positive signal correlating well with published ChIP-seq datasets.
This internship has been an amazing experience for me. I found the work of a researcher both challenging and rewarding and it definitely encouraged me to pursue a career in science. I would recommend this experience to any Biology student who wants to learn how real research is carried forward.
Benedetta Carbone



 (11 votes)
(11 votes)