BSDB Gurdon Summer Studentship Report (17)
Posted by BSDB, on 3 November 2017
![]() Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding.
Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding.
Our sixth report from the 2017 group of student awardees comes from Katarina Grobicki (student at University of Cambridge), who undertook her studentship with Seb Shimeld at the Dept. of Zoology in Oxford.
Hedgehogs & Sea Squirts
In vertebrates, Hh binds to the cell surface receptor Patched, lifting the repression of Patched on Smoothened and allowing signal transduction. In cells not receiving Hh signal, Gli transcription factors are processed to a repressor form (GliR); when Hh signalling is received, this cleavage is inhibited allowing the longer activator form of Gli to persist (GliA). The balance of GliR:GliA is what determines the expression levels of target genes.
In addition to this protein level regulation, signalling also feeds back to regulate the expression of the Gli genes themselves. Ciona are of interest because they are urochordates, so are part of the invertebrate lineage most closely related to vertebrates, which split off before the 2R genome duplication. Vertebrates have 3 Gli genes and their expression is differentially regulated by Hh signalling, whereas Ciona only have 1 Gli gene. I wanted to investigate whether the Ciona Gli gene was also regulated by Hh signalling, or whether this was a novel innovation in vertebrates, made possible by the the 2R duplication.
Making riboprobes
The first step of my project was to synthesise riboprobes for genes involved in the hedgehog pathway (Hh, Ptc, Gli) and a suitable control (FoxIc). Initially I attempted to do this by using PCR with M13F&R primers to amplify the target cDNA from a mini-prepped plasmid, then in vitro transcription to synthesise RNA from this. However after 6 separate PCR and transcription attempts I was still encountering problems. Every PCR of the Gli plasmid produced two DNA fragments (seen by running the products on an agarose gel), even when the primers were swapped (for T7 and T3 primers), a gradient of different temperatures were tried, new PCR reagents were used, and a fresh dilution of the plasmid mini-prep was used. This meant that the Gli PCR product couldn’t be used for probe synthesis. I had successfully amplified the other cDNAs (Ptc, Hh, FoxIc) by PCR however not enough RNA was being produced by the transcription reactions so I decided to restart the probe synthesis from scratch using a different method.
I used restriction enzymes to linearise the plasmids and then transcribed from this linear DNA to successfully produce riboprobes. However the probes were still produced in very small amounts, so I transformed competent cells with my plasmids and grew a mini-prep in order to try again with a much larger initial concentration of DNA. Using the mini-preps I produced large enough concentrations of both positive and negative sense probes for Gli and FoxIc (negative sense probes were used as controls in in situs), and positive sense probe for Ptc, and I purified these using phenol and chloroform.
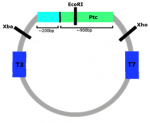
It was not possible to produce as negative sense probe for Ptc due to the Ptc plasmid also containing around 200bp of cDNA encoding part of an RNA-binding protein (this is due to the way the plasmid library has been constructed and this has been missed in previous studies of Ciona Ptc expression). Any riboprobe synthesised using T3 RNA polymerase would also bind to mRNA encoding the widely expressed RNA-binding protein, rendering the probe useless. To produce the positive sense Ptc probe, EcoRI had to be used instead of Xba, so that the DNA encoding the RNA-binding protein was not included.
Once I had produced my probes it was time to collect some Ciona and begin in situ hybridisations.
Ciona work
We drove to a harbour on the south coast to collect gravid adult Ciona which I used to set up in vitro fertilisations. Dissecting Ciona for the fertilisations was tricky and required very steady hands in order to collect eggs without cutting the sperm duct (essential to avoid self-fertilisation). I set up multiple separate fertilisations in the evenings and then incubated these at 17°C overnight, which meant embryos reached early-mid tailbud by the next morning. I then fixed the embryos in paraformaldehyde (PFA) at different ages, ready to use for in situ hybridisations. For earlier stages which had not hatched I had to dechlorionate the embryos before fixing; this must be carried out very rapidly to avoid damaging the embryos.
In situ hybridisations
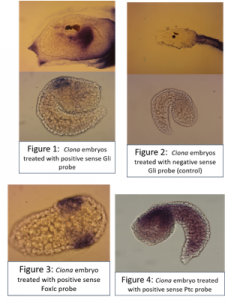
After fixation, embryos were washed in ethanol then prehybridised, before adding 3ul of probe to each eppendorf. The probe was thoroughly washed away and embryos were incubated with blocking solution, then with antibody, then washed many times again before adding the staining solution.Larvae treated with the positive sense Gli probe clearly showed real staining in the head, and tailbud embryos seemed to show staining in neural tissue along their dorsal side, but this was less clear due to background in some of the embryos. In addition, unfertilised eggs were evenly stained all over with the positive sense probe but not stained when treated with the control probe.Embryos treated with the FoxIc probe showed the predicted expression pattern, reassuring me that my protocol was working.My Ptc probe required multiple in situs to optimise the pre hybridisation procedure and find the ideal amount of probe to add. Eventually I was able to see staining towards the back of the head on dorsal side (possibly in ectoderm), but there is still quite a lot of background.
 Cyclopamine experiment
Cyclopamine experiment
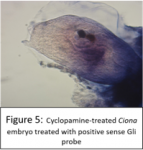
I carried out another set of fertilisations and treated the embryos with cyclopamine (or DMSO as a control) once they reached early tailbud stage; the embryos were fixed in PFA after hatching then used for in situ hybridisations. Cyclopamine inhibits the hedgehog pathway by directly binding to Smo (Chen, J; Taipale, J; Cooper, M; Beachy, P; 2002. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev, Nov 1; 16(21): 2743–2748). I wanted to test how cyclopamine would affect Gli and Ptc expression in Ciona because they only have one Gli gene, which may not be transcriptionally regulated by Hh as it is in vertebrates with their 3 Gli genes. We predicted that Gli expression would not be altered by inhibiting the hedgehog signalling, but Ptc expression would be altered. Unfortunately my embryos stained far too darkly due to me failing to fully remove their outer tunic during prehybridisation, and I ran out of time to repeat this experiment, so I wasn’t able to observe how Ptc and Gli expression patterns were affected.
A few of the cyclopamine-treated embryos with the Gli probe didn’t stain too darkly so expression patterns could still be seen, and these showed the wild-type expression pattern of Gli, which suggests that Gli is not regulated by Hh signalling. However as my Gli controls and all of my Ptc embryos were stained too darkly to see any expression patterns, I can’t conclude anything from these results, as it may have been that the cyclopamine treatment didn’t work. This experiment will hopefully be repeated in the lab soon.
Alternative splicing
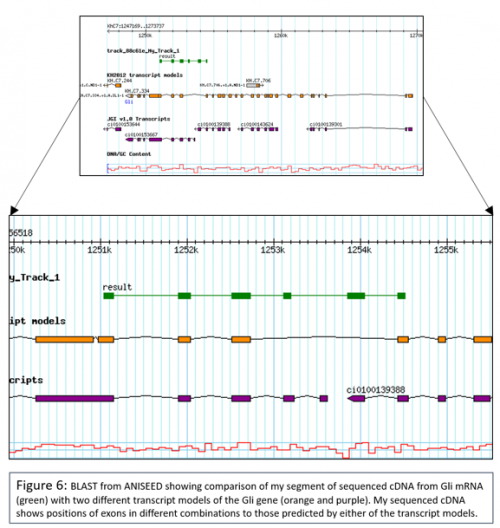
I also briefly investigated whether Ciona Gli might be alternatively spliced, as in Amphioxus. To do this I extracted RNA from Ciona larvae then reverse transcribed from this using oligo(dT) primers to produce cDNA. I carried out a PCR using primers which had been designed to amplify around exons which had been identified as potentially not being included in all transcripts (using the different transcript models available on ANISEED, a very useful integrated database of ascdian genome and expression data). I then ligated my PCR product into a T vector and transformed this into competent cells, which I grew up and then sent for sequencing. My sequence data suggested that Gli isn’t alternatively spliced. My sequence data also suggests that neither the KH2012 transcript model nor the JGIv1.0 transcript model are quite correct in their predictions of exons in Gli, as my cDNA contains 2 exons not predicted by the KH2012 transcript model and doesn’t contain an extra exon predicted by the JGIv1.0 transcript model.
Thank you so much to Seb for hosting me, and to the whole lab for being so welcoming and teaching me so much. Thank you to the BSDB for making it all possible. I really feel like I’ve learned a lot over the summer and I’m now certain I want to pursue a PhD and hopefully a career in research.


 (3 votes)
(3 votes)