BSDB Gurdon Summer Studentship Report (18)
Posted by BSDB, on 30 November 2017
![]() Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding.
Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding.
Our seventh report from the 2017 group of student awardees comes from Agata Czap (student at University College London), who undertook her studentship with Paola Oliveri at University College London.
How to knockout a gene in sea urchin?
I am Agata Czap, an undergraduate student of MSci Human Genetics at UCL. The BDSB Gurdon Summer Studentship has given me an incredible opportunity to undertake research for 8 weeks at Oliveri lab in UCL. I applied to work with Dr. Oliveri to learn about the challenges of applying the CRISPRcas9 technology to new model systems.
My project focused on knocking out the transcription factor Pmar1 in sea urchin S. purpuratus using the CRISPRcas9 system. Pmar1 is a part of a characterized gene regulatory network (GRN) and its role has been dissected using ectopic and dominant negative approaches (1). Specifically, pmar1 represses HesC during early development, as part of a double negative gate (DNG) of skeletogenesis. HesC is another transcription factor, which represses the downstream skeletogenesis genes (2). This DNG enables specification of micromere cell fate in the correct cells (1), from which primary mesenchyme cells (PMCs) will arise and eventually form the larval skeleton. Furthermore, the DNG enables micromeres to release two signals. Early Signal (3), which is responsible for inducing archenteron invagination, and Delta, which activates the Notch receptor in secondary mesenchyme cells for endomesoderm patterning (4) (figure 1).
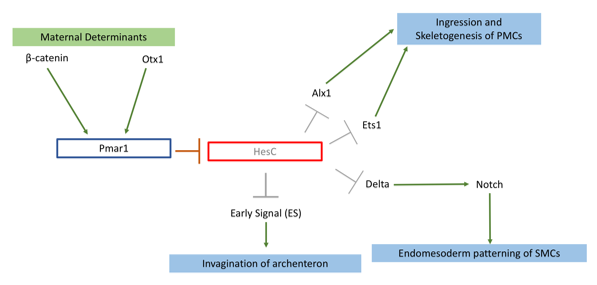
So far, there have been no functional knockouts of Pmar1, as the gene has duplicated 4 times in the S. purpuratus genome. Previous experiments deduced possible function with a series of downregulation and rescue indirect strategies (1,4). Micromeres are located on the vegetal plate of the urchin embryo. One of early experiments carried by Dr. Oliveri, involved removing and transplanting micromeres into animal pole of another healthy embryo (2). This has induced a secondary, ectopic gut (2). Additionally, an embryo from which micromeres have been removed had shown no development of skeleton nor gut (3). Pmar1 expression requires maternal determinant called β-catenin, and imparing its nuclerization results in an embryo similar to micromereless embryo (3). Thus, I have hypothesized that upon successful knockout, the embryo would lack skeleton as well as other endomesoderm derivatives (e.g archenteron). Other ectodermal tissues should be unaffected.
To tackle the issue of pmar1 duplication, I have collected all 4 (Pmar1a-d) FASTA sequences from the Echinobase transcriptome. I have used Jalview to identify conserved regions between duplicates within exon 1 and 2, and generated phylogram to visualize diversification of the sequences. My results agreed with that of Cavaleri’s team (4); Pmar1b has diverged to become a monophyletic group. The sequences are still highly conserved, the first 810 bp of each transcript show 86% similarity. I used this fragment of the Pmar1b sequence, as it had the best consensus when compared to cDNA. I have inserted this sequence into 4 algorithms including CHOPCHOP to generate gRNAs. Unfortunately, the algorithms used different criteria to rate gRNAs. To filter through all 98 gRNAs, I tested each against BLAST in Echinobase for off-targets. This allowed me to obtain top 5 gRNAs, which target every Pmar1 gene.
A more wet lab-based part of my project then followed. During the first two weeks, I have learned how to culture sea urchin embryos, spawn adults to obtain gametes and assess fertilisation rate. S. purpuratus has turned out to be a rewarding model system, embryos can be produced in large quantities and are transparent, allowing clear observation of tissues. Natalie, a PhD student who also worked on applying CRISPRCas9 technology to knockout neuropeptides, has taught me how to synthesize selected gRNAs using PCR and T7 in vitro translation. Once gRNAs were ready, we injected a pair of gRNAs and Cas9 mRNA into oocyte after fertilisation to induce a deletion mutation. The efficiency of knockout was tested with DNA extraction and agarose gels, which measured molecular weight of the target region. I carried out in situ hybridisation and antibody staining on genes acting downstream of Pmar1 to characterise mutants.
My results were promising. In each knockout, I have used PKS1 knockout in pigment cells as positive control, which proved Cas9 enzyme is non-toxic for embryos. PKS1 allows quick visual check that Cas9 works, as albino mutants are easily identified. Furthermore, it has high knockout efficiency of ~90% (5). My knockout attempt had lower PKS1 efficiency with 50% albino incidence at 45hpf. I have used additional positive control, knockout of Jun, a gene downstream of Pmar1, which caused delayed invagination in 42% at 45hpf. Pmar1 gRNA 1 and 5 have been very efficient; at both 45hpf and 70hpf, 100% of embryos were mutants. The embryos did have a blastopore and syncytial rods were present, but the gut nor skeleton were not developed (figure 2).
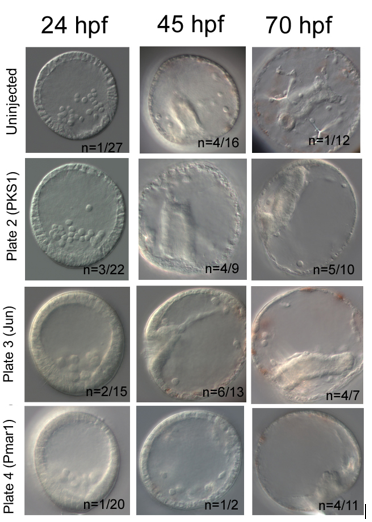
In conclusion, a successful knockout requires a lot of preparation, resilience and knowledge of molecular tools. The procedure gives you greater understanding of how Cas9 enzyme works and of gene function. My gRNAs will be further used by Oliveri lab to test efficiency of Pmar1 knockouts using different combinations of gRNA pairs and Cas9 concentrations.
I would like to thank Dr. Oliveri for giving me a chance to work in her lab. Also to PhD student Natalie Wood for being patient when teaching me all the techniques used. I greatly enjoyed being part of a research team and wish to pursue scientific career further. I would recommend Gurdon studentship to any student applying for a laboratory-based internship.
Bibliography:
- Cavalieri V, Geraci F, Spinelli G. Diversification of spatiotemporal expression and copy number variation of the echinoid hbox12/pmar1/micro1 multigene family. PLOS ONE. 2017;12(3):e0174404.
- Oliveri P, Davidson EH, McClay DR. Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Dev Biol. 2003;258(1):32-43.
- Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proceedings of the National Academy of Sciences. 2008;105(16):5955-62.
- Oulhen N, Wessel GM. Albinism as a visual, in vivo guide for CRISPR/Cas9 functionality in the sea urchin embryo. Mol Reprod Dev. 2016;83(12):1046-7.
- Sethi AJ, Angerer RC, Angerer LM. Gene Regulatory Network Interactions in Sea Urchin Endomesoderm Induction. PLoS Biol. 2009;7(2):e1000029.


 (No Ratings Yet)
(No Ratings Yet)