Canine Somatic Cell Reprogramming and Its Facilitation through Inhibition of Endogenous p53
Posted by Noah Candeli, on 15 November 2021
Numerous efforts have been made to establish bona fide iPSCs from companion animals such dogs and cats. Generation of iPSCs from companion animals would provide useful unrestricted cell resources with a vast scientific potential. To name a few applications, they can be exploited as new models for regenerative medicine and as therapeutic veterinary tools to replace tissues; in veterinary pharmacology for drug development assays, and to elucidate function(s) of genetic variants that are associated with disease.
While protocols for producing human and mouse iPSCs are established, protocols for derivation of iPSCs from domestic animals are slowly developing due to difficulties encountered presumably in their reprogramming process. Only a few studies have indeed focused on the possibility of producing iPSCs from these companion species, and despite some describing their production, the burden of proof is largely lacking.
As an undergraduate student at the University of Edinburgh, this summer despite the current pandemic situation making it harder to find a lab-based studentship, I was lucky enough to have the opportunity to work in Drs. Schoenebeck’s and Burdon’s labs. The Gurdon/BSDB award allowed me to spend two months at Roslin Institute, a pioneering center for genetics and stem cell studies, collaborating with research groups with extensive experience in stem cell research (Dr. Tom Burdon’s) and canine genetics and genomics (Dr. Jeffrey Schoenebeck’s). During my time at Roslin I contributed to the research of an efficient protocol to derive canine iPSCs, supervised by the joint effort of these two excellent lab groups.
Based on the findings that iPSC generation is enhanced by P53 suppression and replacement of L-MYC with C-MYC(Okita et al., 2011) in the set of conventional reprogramming factors (OCT4, SOX2, KLF4, and C-MYC, collectively termed “OSKM” factors); recently, Yoshimatsu et al. (2021) have presented a study which provides insights on the possibility to facilitate canine cell reprogramming. They provided evidence of reprogramming somatic fibroblasts from a canine using an integration-free method. Their 8 episomal (Figure 1) vectors contain the OSKM factors including L-MYC, other pluripotency genes (LIN28 and NANOG), genes that have been shown to facilitate reprogramming (GLIS1and KDM4), and a dominant-negative form of the mouse TRP53 (mP53DD), which was shown to suppress endogenous P53 expression in human cells, and presumably should operate the same in canines.
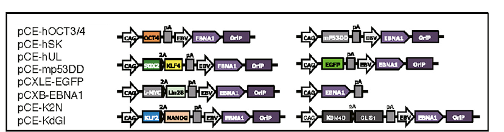
The episomal vectors contain OriP/EBNA1 sequences derived from Epstein-Barr virus (EBV), which ensure the stable extrachromosomal replication of the vectors, hence high expression of the reprogramming factors carried along, which facilitate the production of iPSC. However, the full applicability of this EBV-based system is still unclear as only two dogs were used to prove its actual functionality.
The aim of my project was to assess the ability of the aforementioned system to reprogram canine fibroblasts, testing the capability of facilitating reprogramming by the inclusion of dominant-negative P53. After being introduced to the fundamental cell culturing techniques and practiced such skills on mouse feeder cells, I expanded canine fibroblast from testis in feeder medium prior to transfection of the EBV-based vectors. I then electroporated such cells with 2 different mixtures of vectors, one consisting of the 8 plasmids including the dominant-negative P53 (+mP53DD), and the other without it, consisting of 7 plasmids (-mP3DD). Right after transfection the medium used to feed the cells was changed to M10. Since one of the transfected vectors carried EGFP, I took GFP imaging to directly assess if the transfection was successful, comparing the transfected fibroblast with the non-transfected control (NTC). Images (Figure 2) show a high extent of cell death following electroporation of the cells, while GFP expression in a high proportion of the survived cells indicate uptake of the vectors. Cell recovered and showed prolonged GFP expression until day 14.
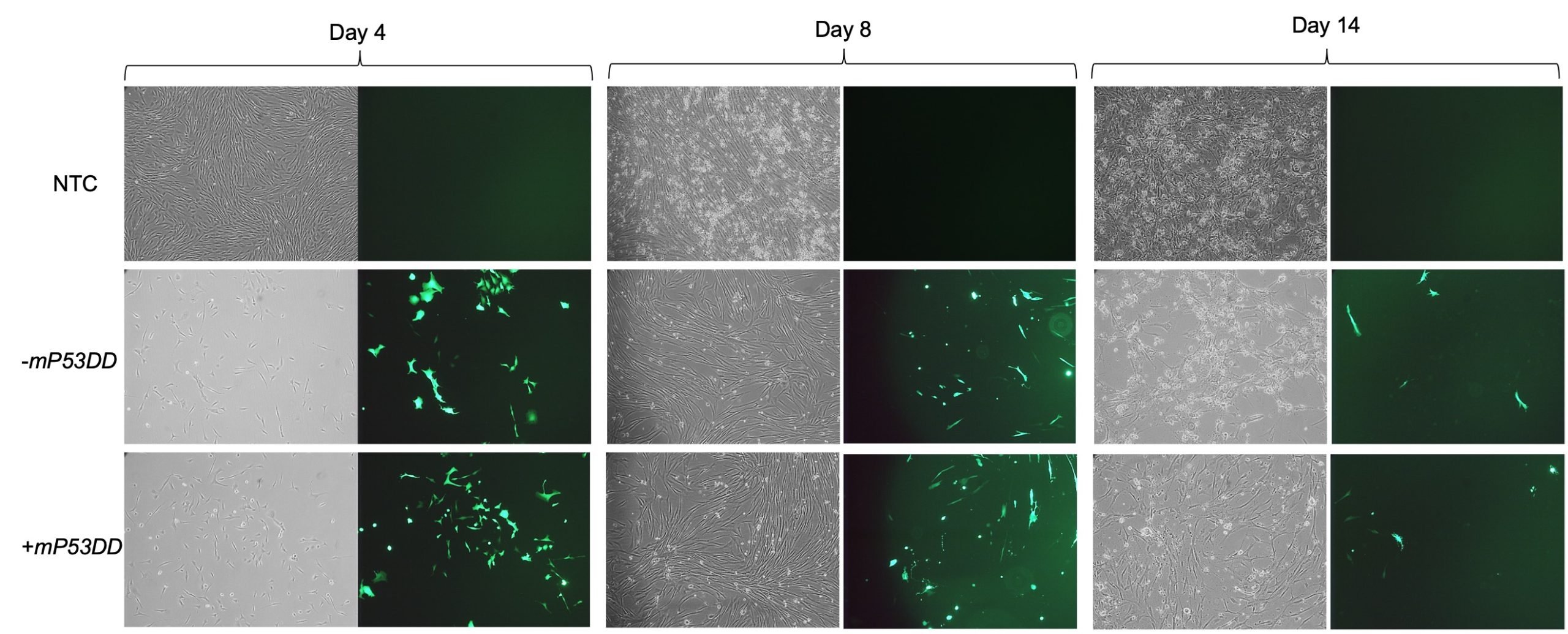
Following pre-expansion for 8 days after transfection, fibroblasts were transferred onto STO feeder cells and changed medium with NSM for induction of iPCS colonies. During reprogramming I sampled cells periodically to harvest their RNA (days 4, 8, and 14). From such RNA samples I obtained cDNAs that I used to perform subsequent RT-qPCR analyses. Using canine specific primers – some of which I personally designed and formerly validated – for endogenous expression of pluripotency markers (POU5F1, NANOG, and SOX2) and other genes of interest (CDH1, CD44, CDNK1A), I was able to assess the reprogramming status of the cells during the process.
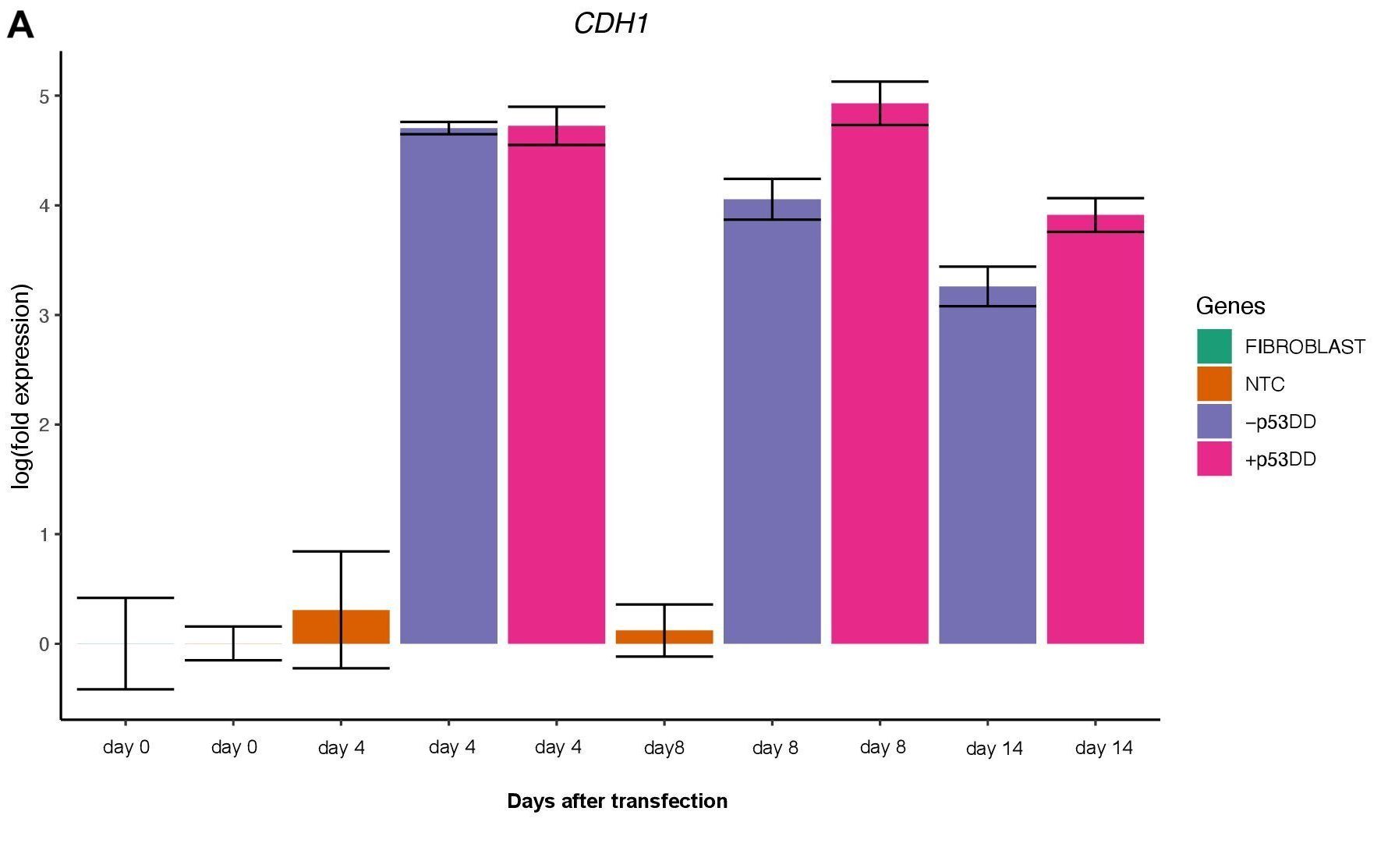
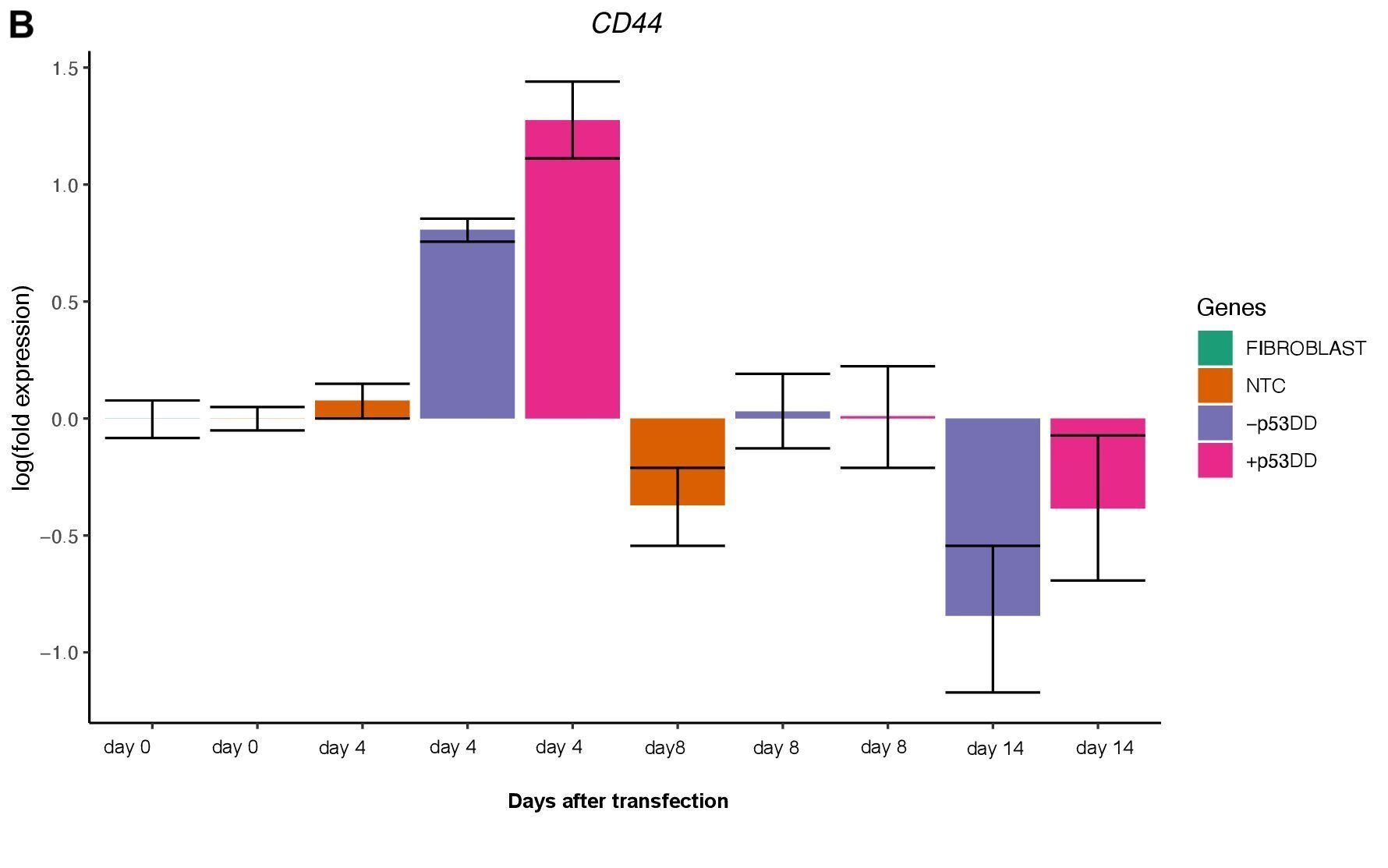
Changes in expression of two markers of reprogramming (CDH1 and CD44) was consistent with what shown in another study (O’Malley et al., 2013). CDH1 (E-cadherin) is upregulated (Figure 3A) indicating mesenchymal-to-epithelial transition which is a typical behavior of the cells entering reprogramming. Furthermore, consistently with O’Malley et al. (2013) CD44 was found to be upregulated at day 4 and progressively downregulated passing the time (Figure 3B), the final population of iPSC are expected to be indeed CD44–.
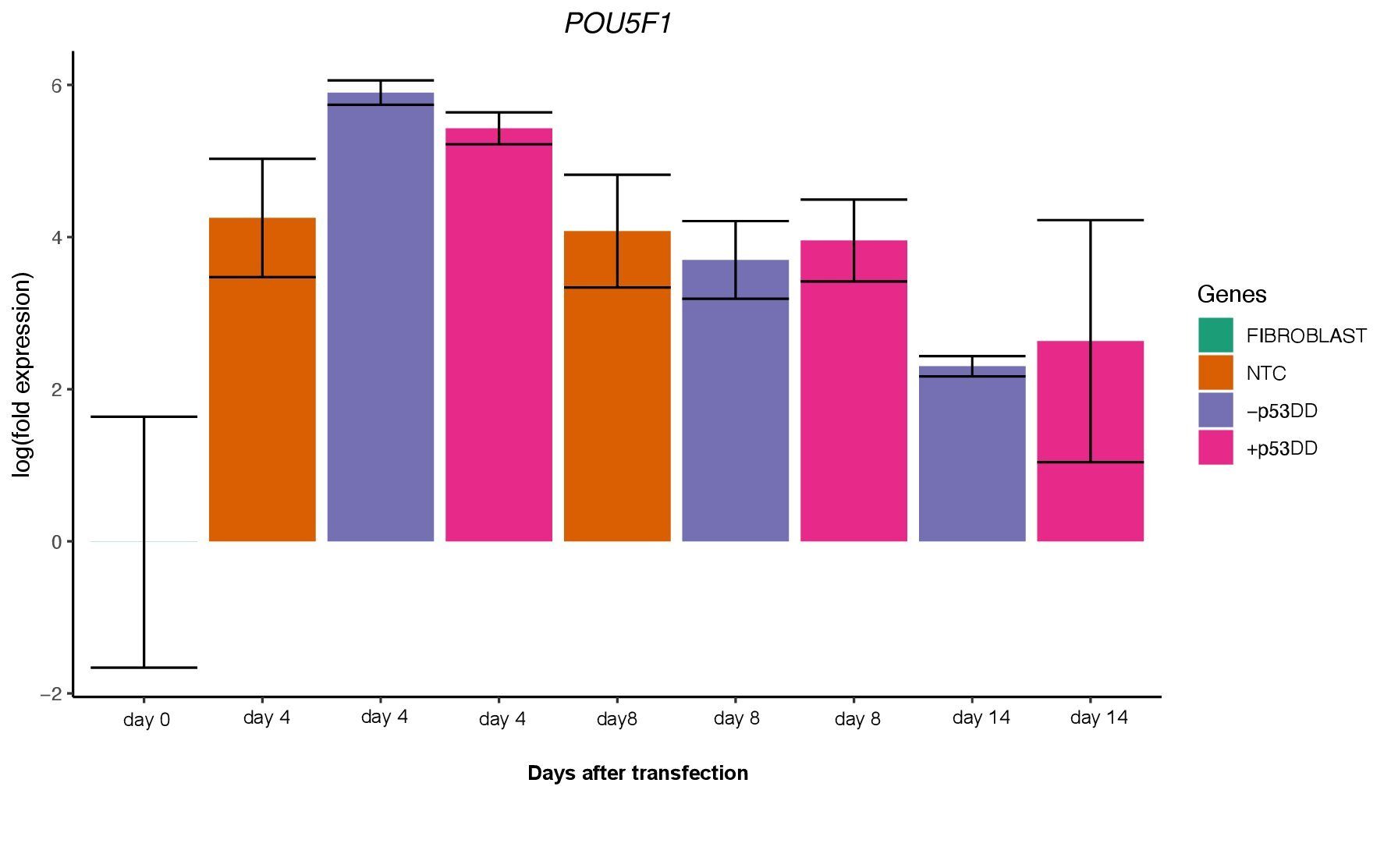
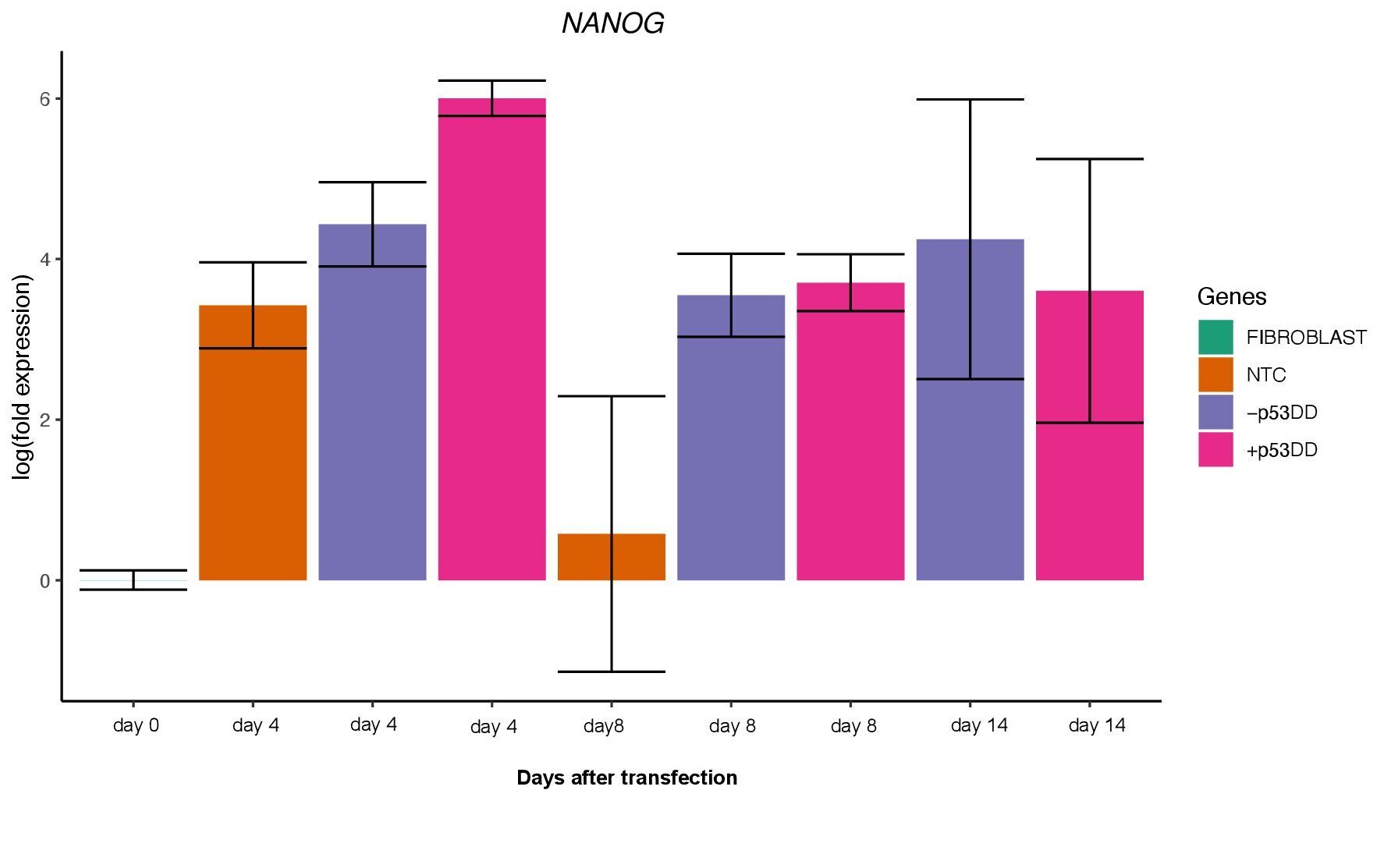
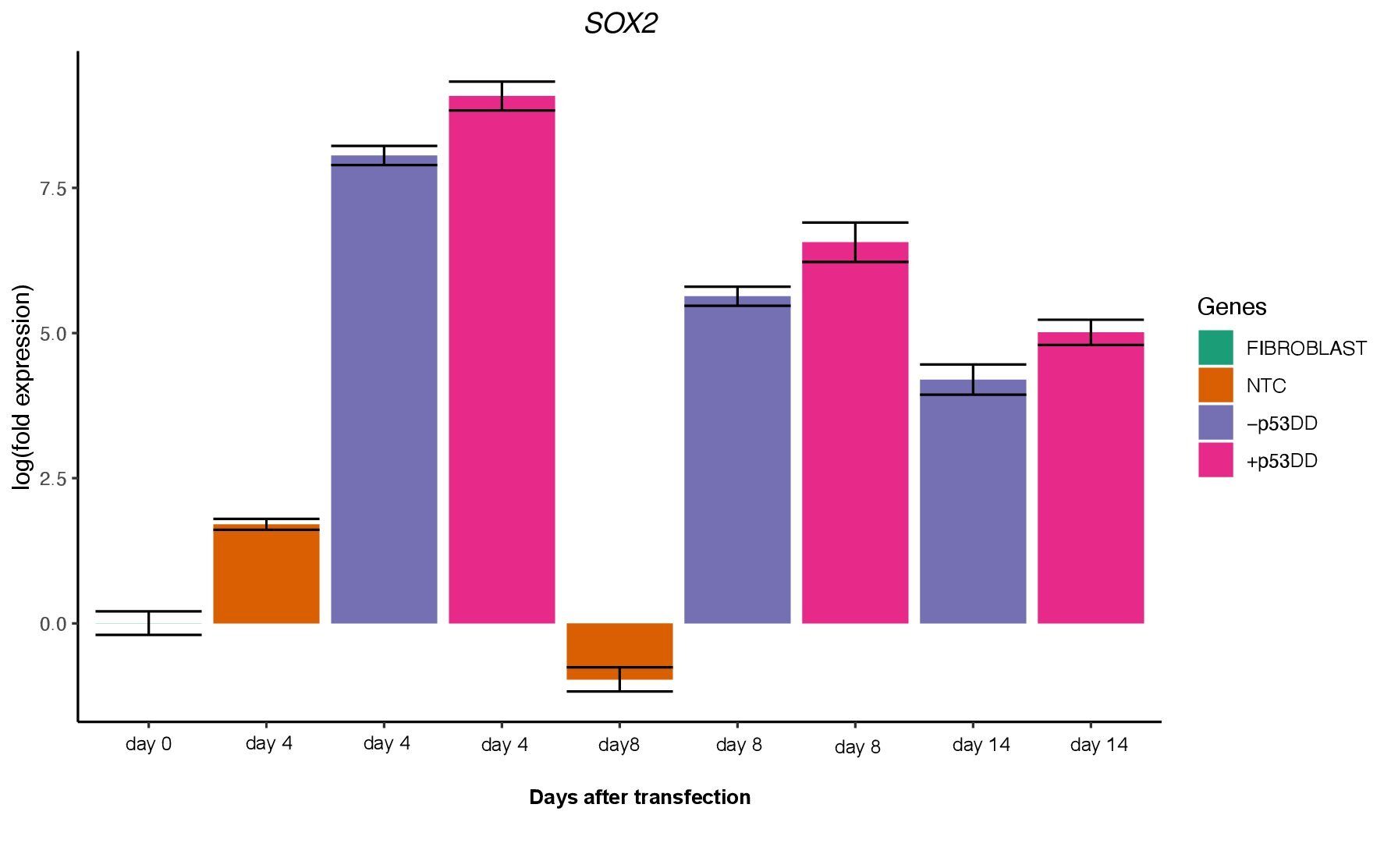
Unfortunately, I was not able to identify any iPSC colony by day 14, as showed in the Yoshimatsu et al. (2021) study, or later in time under either condition (+/- mP53DD). Upregulation of the core pluripotency markers POU5F1, NANOG, and SOX2 during the experiment, demonstrate the ability of the used EBV-based vector system to induce endogenous expression of pluripotency genes in canine cells; however, such expression dissipates throughout time (Figure 4). The reason I could not obtain any iPSC colony might be that this vector system was shown to not maintain sustained enough endogenous expression of the pluripotent genes to overcome the full barrier of reprogramming.
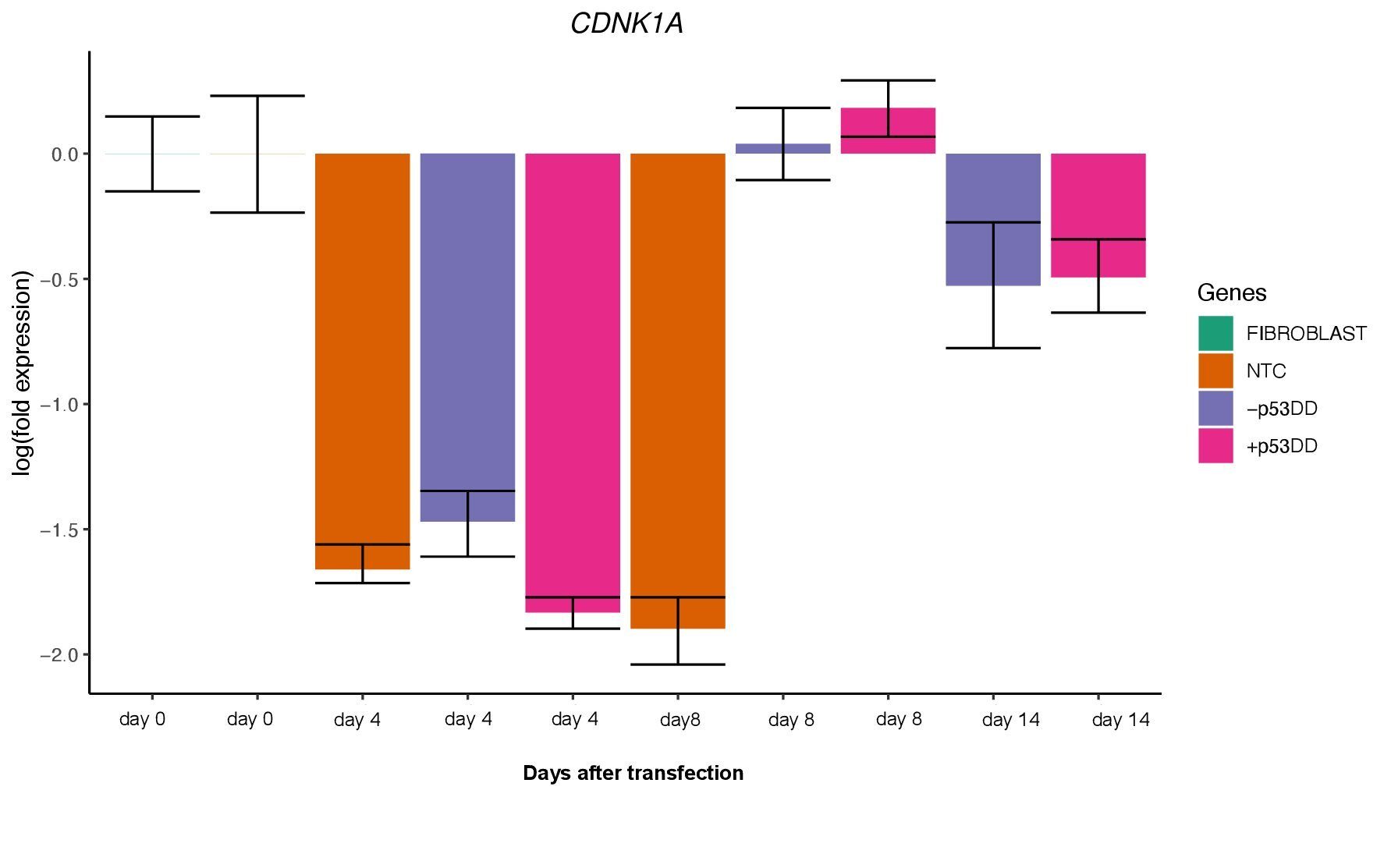
The two vector mixtures +/- mP53DD did not show distinguishable effects, since CDNK1A, direct target of P53showed no difference in expression levels between the two conditions (Figure 5). This suggest that either mP53DDwas expressed at not effective levels or not at all, or that this dominant-negative form of P53 do not interact with the canine form of P53.
Ultimately, this project surely helped to broaden my knowledge in stem cells and reprograming methods, as to learn numerous lab techniques fundamental to pursue hopefully my research career in the future.
References:
- O’Malley, J. et al. (2013) High-resolution analysis with novel cell-surface markers identifies routes to iPS cells.
- Okita, K. et al. (2011) A more efficient method to generate integration-free human iPS cells. Nature methods. [Online] 8 (5), 409–412.
- Yoshimatsu, S. et al. (2021) Non-viral Induction of Transgene-free iPSCs from Somatic Fibroblasts of Multiple Mammalian Species. Stem cell reports. [Online] 16 (4), 754–770.


 (1 votes)
(1 votes)