Colourful life of a fruit fly
Posted by okanca, on 22 January 2014
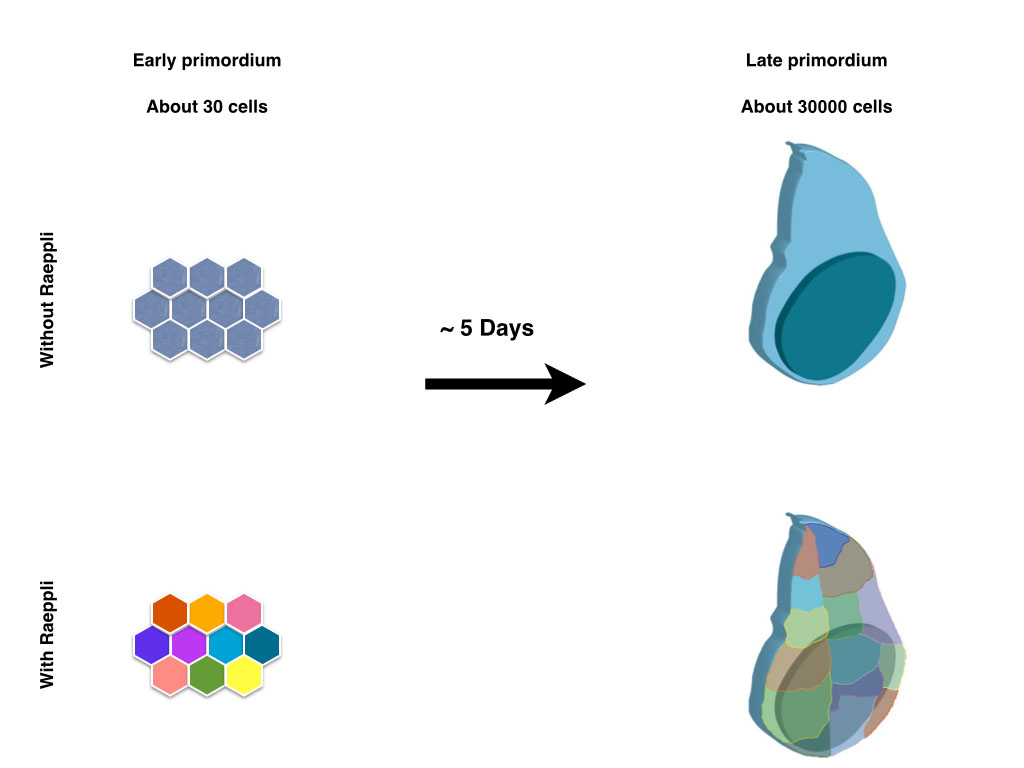
Those lucky scientists, who study organisms which allow live imaging experiments to be effectively performed, do not always appreciate what a luxury it is to watch the tissue of interest develop in time. Scientists who work on less accessible models that take long time to develop to term, look with envy on the beautiful movies of other model systems that show the movement and interaction of the cells, as well as the actions and reactions in the vicinity of a region of interest. Although he developing wing disc in the drosophila larvae serves as a long standing model to study the coordination between tissue growth and patterning, it is not an easy structure to watch as it develops, since it its increase of roughly thousand fold in cell number occurs in a timespan of five days. Larval movement and muscle contractions associated with feeding and other processes make it impossible to image the entire process in vivo and in vitro culture of isolated imaginal discs during the entire growth has not been possible thus far.
In contrast to the limitations with regard to live imaging, the wing disc does serve as an excellent genetic model system. Moreover, since the wing is dispensable for the viability of the fly, a large number of elegant genetic screens allowed to put forward a fine blueprint of its development after decades of research from hundreds of scientists. The larval part of the blueprint that we have gained on wing growth is based on dissecting multiple wing discs of different developmental stages, comparing them to each other and merging the information coming from different discs from the same time point. Using this approach, the information that we can acquire from an individual disc is very limited. This is a limitation that is not restricted to the study of the growth of the wing imaginal disc; other developing organs that take long time to develop suffer from this caveat. Our aim in developing Raeppli was to maximise the information we can extract from each sample (Kanca et al. 2014). We envisioned that marking the lineage of each cells of a young wing disc with a fluorescent protein would allow us to distinguish the contribution of each of those lineages to the adult structure.
We got inspired by the Brainbow and related techniques that are most often used to track the projections of neurones in the developing nervous system (Livet et al. 2007, Hampel et al. 2011, Hadjieconomou et al. 2011) . These techniques use mutually exclusive recombination events to select one of three fluorescent proteins, in individual cells, thereby also marking permanently their progeny. Use of multiple such constructs generates an RGB like colour coding of different cells. We started from this basic idea but diverged from it along the implementation of the idea.
Cre recombinase, which is used for mutually exclusive recombination in Brainbow, is quite toxic in Drosophila, and rarely used in fly research. The Flippase recombinase is somewhat inefficient and causes recombination in a fraction of cells only. We thus opted to use the integrase system, since recombination is both irreversible and efficient. The first versions of constructs we cloned coded for six fluorescent proteins in order to maximise the colour choice per construct. Although these constructs in principle worked to mark cells with different colour combinations, the 6 fluorescent proteins could not be spectrally separated efficiently since there was substantial bleed-through between different colour channels. Although this generates different shades of colours, when two copies of the construct are used the interpretation of the data gets complicated. Thus, for a second version of constructs,we decided to use 4 fluorescent proteins that can be spectrally separated from each other in sequential scans. The fluorescent proteins we chose were selected to be bright and to be usable 1) in combination with GFP, which is applied as a widespread marker, and 2) in combination with far red antibodies. This further increases the amount of information that can be acquired from the sample in addition to the lineage information. We generated two different versions of Raeppli, one where all the fluorescent proteins are directed to membrane by Ras Farnasylation sequence (CAAX), thus marking cell membranes, second where all the fluorescent proteins are directed to nucleus by using Nuclear Localisation Signal (NLS). By using two copies of the construct, we could mark more than 90% of the cells in a wing disc with diverse colour combinations. Moreover, since the fluorescent proteins were bright enough to be visualised through the larval cuticle, we were capable to image the growth of a single wing disc over the three larval periods by acquiring snapshots of the disc from an anaesthetised larva. Multiple individually marked clones served as reference points to compare the different time points. Moreover, we opted to increase the flexibility of usage of Raeppli by including a cassette that, when unrecombined, can respond to either Gal4 or LexA, and when recombined by Cre, can respond to either one or the other binary system. Thus, for routine uses one can express the proteins with any driver that one wants, but for more specific uses, such as over-expression studies using an additional transgene, in combination to lineage analysis, one can use one binary system for detection of Raeppli and the other for manipulating the cells.
Although the analyses shown in the paper are confined the study of wild type tissue behaviour, we are planning to explore the flexibility and efficacy of Raeppli and analyse the whole tissue response induced by perturbations in parts of the tissue. Due to the flexibility of Raeppli, we believe that its use will help to address numerous interesting scientific questions, both at the cellular as well as at the tissue level. Finally, together with recent advances in cell labeling tools, Raeppli will help to shed new light on organogenesis (Boulina et al. 2013, Wortley et al. 2013, Kanca et al. 2014) .
References:
1. Boulina, M., Samarajeewa, H., Baker, J. D., Kim, M. D. & Chiba, A. Live imaging of multicolor-labeled cells in Drosophila. Development 140, 1605–1613 (2013).
2. Hadjieconomou, D. et al. Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nat. Methods 8, 260–266 (2011).
3. Hampel, S. et al. Drosophila Brainbow: a recombinase-based fluorescence labeling technique to subdivide neural expression patterns. Nat. Methods 8, 253–259 (2011).
4. Kanca, O., Caussinus, E., Denes, A. S., Percival-Smith, A. & Affolter, M. Raeppli: a whole-tissue labeling tool for live imaging of Drosophila development. Development 141, 472–480 (2014).
5. Livet, J. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007).
6. Worley, M. I., Setiawan, L. & Hariharan, I. K. TIE-DYE: a combinatorial marking system to visualize and genetically manipulate clones during development in Drosophila melanogaster. Development 140, 3275–3284 (2013).


 (9 votes)
(9 votes)
If you are also interested in the more artistic side of science you may want to check Development’s cover featuring a very colourful Drosophila pupa generated using this technique: http://dev.biologists.org/content/141/2.cover-expansion