Counting transcripts to track cell state
Posted by Martin Jakt, on 16 December 2012
Many years ago, we started to use micro-arrays to look at how gene expression changes during differentiation of lateral mesoderm. In particular we were interested in differentiation leading to the endothelial and hematopoietic lineages (derivatives of lateral mesoderm) and we performed array experiments on populations of cells sorted by surface molecules at different stages of the process. Although these identified Etv2 as the primary driver of the primitive to lateral mesoderm transition (Kataoka 2011), as well as pretty much all genes induced as a result, we were unable to draw any conclusions as to the nature of the underlying system that controls this process. Even simple observations which on the surface have obvious explanations could be interpreted as being evidence of a range of phenomena. For example, we observed apparent co-expression at low levels of genes associated with both erythropoiesis and endothelial identities in lateral mesoderm; this is kind of expected, but in truth, we cannot even conclude to have seen that, as the genes may not be co-expressed in the same cells, nor can we actually state that the expression is low, as it may simply be observed in a small fraction of the cells. Similarly we observed oddities like hemoglobin gene expression apparently preceding the expression of it’s presumed activator Gata1, which on the surface seems interesting, but which is most likely an artefact due to differences in promoter strengths combined with cellular heterogeneity.
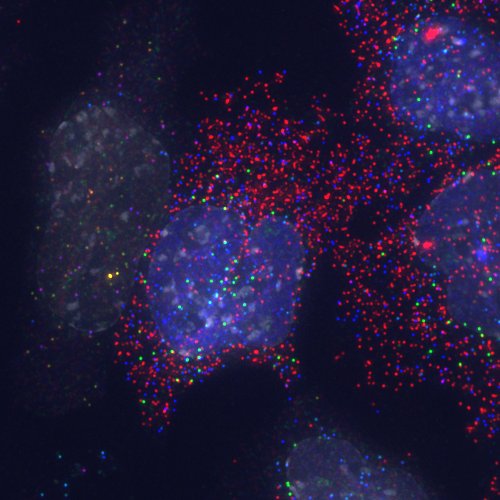
Detection of transcripts by candy FISH and an EGFP-Etv2 fusion protein by direct fluorescence. Lateral mesoderm differentiation was induced by expression of an EGFP-Etv2 fusion protein and transcripts detected by candy FISH. Transcripts: Fli1 (blue), Cdh5 (green), Flk1 (blue + green -> cyan), Etv2 (red), Pdgfra (blue + red -> purple), Snail1 (green + red -> yellow). The EGFP-Etv2 fusion protein can be seen as an even nuclear signal (blue) in most of the nuclei and sites of transcription appear as intense nuclear signals. DAPI (white) indicates nuclei. Pseudocolors: Alexa 488 and EGFP blue; Cy3 green; Cy5 Red; DAPI white.
This led us to search for some means of estimating gene expression within single cells; as we wanted to detect co-expression of genes, whatever method used needed to allow measurements of expression from at least two genes, but since co -induction or -expression may occur in different cell states specifically, the more genes we could observe simultaneously the better. We also strongly wanted a method which would provide some way of judging the accuracy of any measurements as it would otherwise be very difficult to interpret low frequency events.
It had already been shown in 2002 that combinatorial fluoresecent in situ hybridisation (FISH) can be used to detect sites of transcription from up to ten genes simultaneously (Levsky 2002). Combinatorial detection, or encoding, of identities relies on the ability of spatially segregating individual sites containing signals and since it had been shown even earlier that FISH combined with high-resolution microscopy makes it possible to detect single transcripts (Femino 1998), it had been obvious for some time that the combination of these two methods ought to allow the enumeration of transcripts in a combinatorial fashion. However, reliably detecting single transcripts is more difficult than detecting sites of transcription (which usually contain many copies of the transcript) and we made use of an improved protocol (Raj 2008) that uses large numbers (~48) of weakly labelled probes targeted to individual transcripts. This provides sequence dependent signal amplification, and we used this to demonstrate the reliable detection of transcripts using specific combinations of fluorophores for each transcript (Jakt 2013). Given the complexity of the hybridisation (simultaneous use of 100s of probes) it is somewhat surprising quite how well it works; the resulting colours show a great range and vibrancy reminding us of an assortment of candies (artificially coloured no doubt) leading us to propose the term candy FISH.
In routine use we have been able to use only three fluorophores for detecting transcripts, and this limits us to a maximum of 7 genes; however, the methodology should extend easily to 10 genes or more depending on the number of usable fluorophores, the resolution of the microscopy and the level of expression of genes. Indeed, recently Lubeck et al. (2012) demonstrated the detection of transcripts from 32 genes simultaneously using super-resolution microscopy based upon switchable fluorophores and statistical imaging (STORM).
We used candy FISH to analyse gene expression of a number of genes associated with vascular and blood differentiation (Etv2, Tal1, Fli1, Gata2, Runx1 and Cdh5) during differentiation of ES derived mesoderm cells. Initially we had been concerned primarily with determining the extent of heterogeneity within differentiating cells and using such information to refine analyses of micro-array gene expression data. However, the data itself has properties that reveal much more than we had initially considered. Since descriptive power increases exponentially with parameter number, a limited number of genes can describe a wide range of cell states, and the data can be used to visualise the set of cell states that appear during differentiation. In our analysis we were able to visualise a continuum of identities corresponding to stage of differentiation from cells at a single time-point. Somewhat surprisingly, cells within the endothelial lineage essentially co-expressed all genes assayed with levels varying along the primary axis of differentiation in a coordinated manner, suggesting that maturation along this axis is a largely deterministic process. In contrast, the timing of expression of Etv2 (which is necessary for lineage entry) appeared largely stochastic, suggesting different mechanisms for lineage entry and maturation.
Currently most effort expended towards explaining mechanisms governing biological phenomena is focused on identifying gene interactions and from there deducing gene regulatory networks. Such networks often appear to have explanatory power, but it is difficult to determine both appropriate functions and parameters that recapitulate the biological systems. Etv2 has been proposed to act through the three transcription factors Gata2, Tal1 and Fli1, which in turn are thought to be able to form a positively reinforcing triad motif that stabilises the state of hematopoietic precursors (Pimanda 2007). In our data we see a strong correlation in expression between these three factors suggesting that such a network might be operating; however, superimposing the axis of differentiation on our data indicated that the expression of these factors is lost during endothelial maturation and that their correlation in expression is more likely related to commonality in upstream regulation, and that for unknown reasons the triad motif fails to engage during this process. In this case there are clearly many unknown gene interactions that drive the process, but this example highlights the difficulty of modelling gene regulatory networks in the absence of cellular data.
The use of FISH to enumerate transcripts has several advantages over more commonly used means of estimating gene expression at the single cell level. In particular the measurements are absolute numbers of transcripts making it trivial to compare levels across different genes and samples. Perhaps more importantly, the measurements are made in situ and hence allow the affects of cellular interactions to be assessed. In addition the method is compatible with antibody staining and as such allows the simultaneous detection of protein and transcripts. This should also allow it to be combined with methods like in situ proximity ligation in order to also assess the state of signalling cascades and how signalling drives gene expression.
The future brings with it hopes of understanding complex biological phenomena such as embryonic differentiation through computational modelling of the interactions between regulators and regulatees. Such models make predictions of cellular behaviour, which in the case of differentiation of multipotent cells must include the generation of diversity. Methods such as candy FISH allow not only the direct observation of the behaviour of systems at the individual cell level, but also make it possible to take into account effects of interactions between cells thus turning the problem on its head. We believe that this is crucial for the development of credible models of differentiation, and that when used in combination with more classical approaches will eventually provide the ability to model complex cellular behaviour. In the meanwhile, the simple scaling up of the analysis to larger numbers of cells will provide an abundance of numbers that are intrinsically linked to the basic manner in which genes are regulated.
Jakt L.M., Moriwaki S. & Nishikawa S. (2013). A continuum of transcriptional identities visualized by combinatorial fluorescent in situ hybridization, Development, 140 (1) 216-225. DOI: 10.1242/dev.086975


 (2 votes)
(2 votes)