Creative morphometrics – so many tools, so little time
Posted by Milos Blagojevic, on 4 August 2014
Nowadays, the hardest thing in science is similar to what we experience in daily life, that is organization and choice. In a virtual plethora of techniques, methods and analyses, an aspiring researcher is faced with a flood of information, achievements and tools of the trade. Especially so with computers. But if it wasn’t for this ongoing torrent, some great synthetic approaches in studying biological variation would not emerge. Geometric morphometrics is an example of one such approach, as it greatly benefited from the ever increasing informational content all around biology, mathematics, statistics and computer science. If one is exposed to this multitudinous information, then organizational skills must take over natural curiosity, which is not very likely. Another important choice to be made regards the way creativity is shaped, the way it is displayed for others to see. This choice presents a balance between organization and chaos, a natural creativity border. In the blog Creative Morphometrics I choose some tools, R, python and octopress blogging platform, to try to tell one small part of the story about the wonderful world of biological shape analysis, its visualizations and the ways it connects to underlining phenomena, such as ontogeny.
For now, not much underlining and outward connections has been made in the blog, but its concept will be adapted over time (hopefully it is adaptable, at least in the design). Each post mostly deals with one particular idea, a problem in shape analysis and its proposed solution, along with results and R/python code. Of course, the emphasis, for now is put to techniques and tools (R or python packages) which can produce results that are both visually appealing and informative. Python-related posts are more basic (as my knowledge of python) and are there to “bring” geometric morphometrics to python, since there are no readily available packages for these analyses. R posts are the creative part (debatable creativity), with some not so common tasks in morphometrics, but also in data analysis in general.
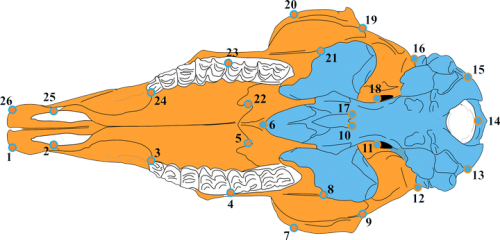
One common point between quantification of morphological variation and its underlining developmental interactions, especially in focus within the concept of evo-devo, is modularity in the complex morphological structures. All morphometric analyses are underpinned by modularity, as it is one of the most important determinants of variability of both parts and the hole of adult morphology. The data I was working on during my PhD thesis research, mammalian crania (Figure below), exhibited constant and complex pattern of modularity across populations, and future posts in Creative morphometrics will surely include more aspects of modularity, as well as techniques that will enable the a-posteriori (purely analytical) determination of modules. Modularity represents the pattern of covariation between parts that share common developmental origin or form the same functional unit. Most informative papers about this key concept are “Evolution of covariance in the mammalian skull” by Hallgrimmson et al., “Morphological integration and developmental modularity” by Klingenberg and “The road to modularity” by Wagner et al. They bring closer the observed phenotypic variability and the developmental interactions that can both produce and maintain patterns of covariance in the complex morphological structures, enabling better understanding of the interactions between development, morphology and the environment.
Developmental modules of the chamois goat skull (orange – predominantly neural crest, blue – predominantly somitomeric origin of the cranial elements).
In conclusion to this post, the title can be repeated, as, with R, python and tremendous computational and analytic power available today our natural scientific curiosity can only sigh that there are so many tools and so little time. Researchers must make choices and only through communication with others will these choices flourish and bring forth the results.


 (1 votes)
(1 votes)