Dynamic new roles for local RNA regulation in neural stem cells of the developing brain
Posted by Debra Silver, on 22 December 2016
Debby Silver and Louis-Jan Pilaz
Comment on Pilaz, at al. Current Biology. 26(24): 3383-3392
Neurons and glia of the developing brain are produced from an elegant cell cell type called radial glia. These stem cells are fascinating not only because of their inherent multipotent nature, but also because of their unique bipolar morphology. Radial glia are polarized, with a cell body near the ventricle and extending from this a long basal process which forms endfeet at the top of the brain. This basal process can range in length from 50 microns to over 1000 microns long in human brains! Although these basal structures are known to be essential for controlling neuron generation, very little is known about their molecular regulation.
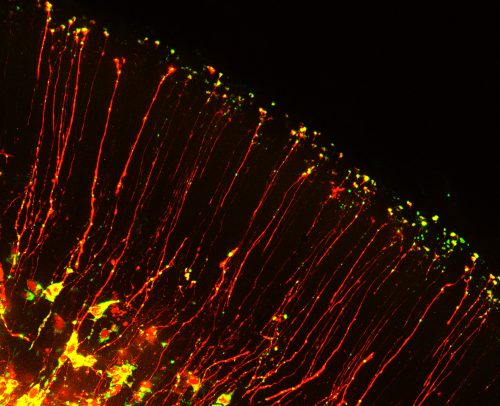
The beautiful morphology of radial glia, which are reminiscent of post-mitotic neurons with growing axons, inspired us to think about roles for mRNA localization in neural stem cells. In post-mitotic neurons local translation is relevant for synaptic function, axon guidance and wound healing. But could RNA be transported with basal processes and locally translated in the distal structures of neural stem cells? This was a natural question for us to ask, given that a major focus of our lab is to understand how RNA regulation impacts neural stem cell function. A handful of studies, including recent studies from Osumi and colleagues1, had identified several RNAs localized to endfeet. Yet it was unknown if neural stem cells contain a local, unique transcriptome. Moreover, it was mysterious whether these mRNAs reached endfeet by active or passive movements. We were excited to combine our interests in RNA biology with knowledge of live imaging and brain development to investigate these questions. In doing so we were faced with three major challenges.
The first challenge we faced was to image RNA movements in an intact tissue. As far as we know, this had never been accomplished in a mammalian model. One of the ways to visualize RNAs by live imaging is with the MS2 system, which requires expression of two types of molecules2,3. The RNA of interest includes MS2 RNA loops (we used 24 loops for our reporters). This RNA is expressed along with a GFP-fused MS2 coat protein, which has nuclear localization signals and high affinity for the MS2 loops. Using this system Singer and colleagues were able to image actin mRNAs transcribed in a mammalian brain4. To visualize RNA transport in brain slices, we used in utero electroporation to express the 3’UTR of the Ccnd2 RNA as a reporter, which was shown by Osumi and colleagues to accumulate in radial glia endfeet1. Then, we had to really push the limits of live imaging embryonic brain slices. RNAs move fast. This involved a lot of late night live imaging sessions, not being sure of what kind of signal to look for. But then one night, there they were! We could actually see bright dots moving through the eyepiece; it was amazing. In the end we had to image the samples with the laser set on high power, which means that some photobleaching occurred and the samples could only be imaged for one or two minutes at a time. After systematic analyses of many basal processes, we realized the moving RNAs may have originally been hard to find because it was only evident in one third of the neural stem cells. After this realization and persistence, we extended our studies to image RNAs moving in radial glia at different stages of development, and observed that their speeds and processivity (run lengths) changed across developmental time. This established that RNAs actively move in neural stem cells!
The second challenge we faced was to image local translation in radial glia endfeet. We decided the best approach for imaging was to use photoconvertible proteins, which have been used extensively by Christine Holt and colleagues5. We expressed a reporter construct expressing a photoconvertible protein coupled to an mRNA localization element (Ccnd2 3’ UTR in our case). After photoconversion of the protein from green to red using UV light, the recovery of the natively green fluorescent protein could be examined over time. One of the best controls for these experiments is to use a reporter lacking the RNA localization signal. In our first attempts, we tried to image endfeet within intact brain slices. In control conditions, one expects to observe little to no recovery, since no RNA is localized to the endfeet. However, we could still observe significant recovery in this condition, which we attributed to diffusion of newly synthesized proteins from the radial glia cell body and basal process. This was a hurdle and we were not sure how to proceed…until one day, while making organotypic brain slices containing radial glial cells filled with fluorescent GFP, we noticed a piece of tissue floating away from the slice that contains fluorescent “particles”. After careful characterization, it turned out that this piece of tissue contained the basement membrane, the overlying meningeal cells and GFP-positive basal endfeet severed from radial glia basal processes! We then realized that by peeling the meninges/basement membrane off of the brain, we had found a way to isolate radial glia basal endfeet and measure translation. We were now able to culture those isolated endfeet and the surrounding tissue to perform local translation experiments without too much trouble. This showed that indeed, some mRNAs are capable of local translation within neural stem cells, and also suggested this could be a regulated process!
Finally, the third major challenge we were faced was to find a way to identify basal endfeet RNAs en masse. We first contemplated using laser capture microscopy to isolate basal endfeet from a tissue. This would have been a very daunting task given the size of the endfeet, and the amount of RNA necessary to perform downstream analyses. However, thanks to our discovery that basal endfeet could be easily isolated from radial glia, we had an path to solve this problem. Although these endfeet preparations contain other cells including neurons, vasculature, and fibroblast from the meninges, we found a way to specifically isolate RNAs from endfeet. We adopted an RNA immunoprecipitation approach pioneered by Jack Keene and colleagues6. For our purposes we used in utero electroporation to introduce a tagged version of an RNA binding protein into radial glia, and then isolated endfeet from transfected cells. We used EGFP-FMRP because we had already found this protein strongly accumulates in basal endfeet. This strategy enabled us to identify 115 RNAs that localize to basal endfeet. Validations using RNA FISH showed that the RNAs we pulled down were bona fide residents of radial glia endfeet. This discovery uncovered a local transcriptome in nerual stem cells, enriched for microtubule and signaling associated proteins!
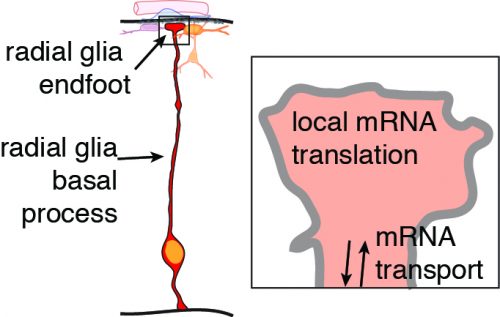
We are very excited about the discovery that mRNA can actively move and be locally translated. By overcoming these technical challenges, this study taught us that radial glia serve as highways for molecular transport, not only of mRNAs but also proteins, including RNA binding proteins. There are many questions we are now working to address. What other types of molecules are rapidly transported in radial glia and when does this occur? What other RNAs are located in radial glia endfeet? What controls their transport and translation? And perhaps the most important and challenging question, what is the function of local translation? We are excited for the new lessons we will learn as we peer inside radial glia.
Find out more about work in the Silver Lab at https://sites.duke.edu/silverlab/
References
1 – Tsunekawa, Y., Britto, J.M., Takahashi, M., Polleux, F., Tan, S.-S., and Osumi, N. (2012). Cyclin D2 in the basal process of neural progenitors is linked to non-equivalent cell fates. EMBO J. 31, 1879–1892.
2 – Buxbaum, A.R., Haimovich, G., and Singer, R.H. (2015). In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 16, 95–109.
3 – Bertrand, E., Chartrand, P., Schaefer, M., Shenoy, S.M., Singer, R.H., and Long, R.M. (1998). Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445.
4 – Park, H.Y., Lim, H., Yoon, Y.J., Follenzi, A., Nwokafor, C., Lopez-Jones, M., Meng, X., and Singer, R.H. (2014). Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science 343, 422–424.
5 – Leung, K.-M., van Horck, F.P.G., Lin, A.C., Allison, R., Standart, N., and Holt, C.E. (2006). Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat. Neurosci. 9, 1247– 1256.
6 – Keene, J.D., Komisarow, J.M., and Friedersdorf, M.B. (2006). RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 1, 302–307.


 (6 votes)
(6 votes)