Featured resource: Human Developmental Biology Resource (HDBR) Atlas
Posted by the Node, on 13 June 2022
Doing great science depends on teamwork, whether this is within the lab or in collaboration with other labs. However, sometimes the resources that support our work can be overlooked. Our ‘Featured resource’ series aims to shine a light on these unsung heroes of the science world. In our latest article, we hear from Steve Lisgo and Janet Kerwin, who describe the work of the Human Developmental Biology Resource (HDBR) Atlas.
When was the HDBR Atlas established and what are its aims?
The HDBR Atlas was established in 2020 and was developed from the Human Developmental Studies Network (HuDSeN) and Electronic Atlas of the Developing Human Brain (EADHB) projects, which began in 2002. The HDBR Atlas brings together 3D models, high resolution histology sections and gene expression data, and aims to facilitate the learning and understanding of human embryonic and fetal development.
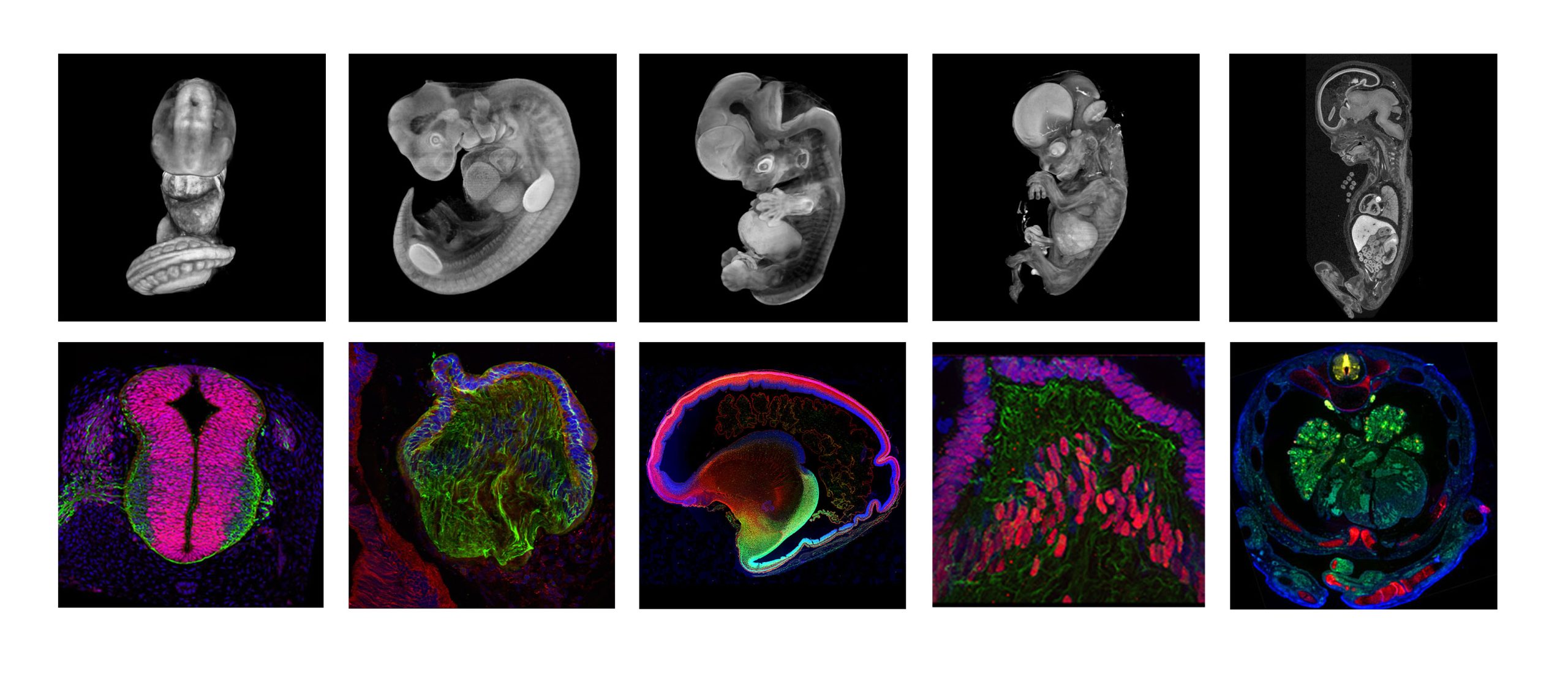
Who runs the HDBR Atlas?
The Human Developmental Biology Resource (HDBR; www.hdbr.org) is an MRC/Wellcome funded human embryonic and fetal tissue bank, managed across two centres: Newcastle University and UCL. The samples collected by the resource are used in registered research projects in the UK and overseas. Data generated both by HDBR users and produced in house at London and Newcastle HDBR centres are disseminated through the HDBR Atlas. The Newcastle HDBR group lead on curating and developing the website.
What can researchers find in the HDBR Atlas?
The HDBR Atlas is a freely available website comprising four sections: 1) 3D models of normal embryos (Carnegie stages (CS)12 to CS23) and fetuses (8-11-post conception weeks), plus a series of karyotypically abnormal embryos. 2) Annotated histology sections of whole CS12-23 embryos at low and high resolution. 3) Organ systems, including gastrointestinal, nervous, urogenital and visual systems, and with a new feature on the developing cardiovascular system. The latter includes movies and interactive 3D models (CS12-16), annotated H&E sections (CS14-22), and fetal heart dissections (8-17 post conception weeks (pcw)). 4) Gene expression data for 129 genes from CS11 to 19 pcw. All the HDBR Atlas resources are available to download on request.
How can the community contribute?
We encourage users of HDBR material to return data images (or physical slides, if this is easier) so that the data can be made publicly available via the HDBR Atlas. Anyone who has human embryonic or fetal protein or gene expression data generated from microscope slides can submit data to us using the submission form available on our website (https://hdbratlas.org/data-submission.html). Expertise in different organ systems is always welcome to define anatomy in the 3D models and annotate 2D histology sections.
Any hidden gems, features that are new, or that researchers might be less aware of?
Previously, we have shown 2D images and movies of 3D models on the Atlas. Recently, we have added interactive 3D reconstructions of early heart development (https://hdbratlas.org/interactive/heart.html) which can be manipulated and zoomed in/out by the user.
If money was no object, what would you like to add to the HDBR Atlas?
Generate 3D models for all stages of development from 3 to 20pcw; create more detailed/comprehensive labelling of the histology sections; further develop the high resolution eHistology viewer to include transverse, sagittal and frontal section planes at each Carnegie Stage, and expand this into fetal stages; produce a fully searchable gene expression database with sets of marker genes for key structures e.g. cardiac markers and distinct brain regions; and create a section on comparisons between human and mouse development for different organ systems.
Feedback from users of the Atlas
‘Many thanks for a wonderful resource!’
‘I would like to take this opportunity to reiterate how useful the HDBR is. It is an amazing resource, unequal in its ability to support developmental biology research. My sincere gratitude to all the dedicated staff involved.’
‘I have used the 3D images in lectures (thanks!) and would really like to see these accompany any further additions/stages. 2D histology sections are also extremely useful to help lab members understand what they should be looking for in our own experiments.’
‘This is a great project! We linked to certain images for our graduate embryology workshops last semester. The students really liked it!’
‘Again, thank you for putting together this incredible resource.’
Thanks to everyone who has contributed to our ‘Featured resource’ series so far. You can read the whole series here. Let us know which resource(s) is essential for your research and we’ll add them to our list. Get in touch at thenode@biologists.com.


 (4 votes)
(4 votes)