Featured Resource: MorphoNet
Posted by the Node, on 10 January 2023
Doing great science depends on teamwork, whether this is within the lab or in collaboration with other labs. However, sometimes the resources that support our work can be overlooked. Our ‘Featured resource’ series aims to shine a light on these unsung heroes of the science world. In our latest article, we hear from Patrick Lemaire and Emmanuel Faure, who describes the work of MorphoNet.
Patrick Lemaire1 and Emmanuel Faure 2
- CRBM, Université de Montpellier, CNRS, 34293 Montpellier, France.
- LIRMM, Université de Montpellier, CNRS, 34095 Montpellier, France
What is MorphoNet?
MorphoNet (1) is an interactive morphodynamic web browser designed to help scientists, teachers and students share, analyze and visualize the large 3D morphological datasets that can be generated by modern imaging technology, ranging from live light sheet microscopy of cells and embryos to X Ray tomography of fossils.
Why did you build MorphoNet?
We initially created MorphoNet so that our biologist collaborators can interact, without programming skills, with the 3D + time datasets they obtained by light sheet microscopy of live ascidian embryos followed by segmentation (2). We wanted the system to be web-based so that they do not have to install any sophisticated software on their computers, and to allow data sharing. We therefore chose to use a very popular gaming engine Unity, to show meshed segmented data. This choice allowed us to compress the complex 3D + time datasets sufficiently so that they could be explored, curated and shared through an intuitive graphical user interface, running on a standard web browser. In a similar way as genomic browsers display genetic features and epigenetic or gene expression data as traces onto the primary genome sequence, quantitative and qualitative information can be imported and projected onto individual or grouped segmented objects in MorphoNet. These ‘morphological augmentations’ can be saved and shared with other users (and used in publications), respecting the FAIR philosophy.
Can I use the system with my data?
Of course! It’s open-source, publicly available and we commit not to make any scientific use of your private data! After a free registration to the service, you can upload up to 10Gb of data. This space is sufficient for 20 movies like the one shown in Figure 1 – top left. If you need more storage, please contact us, we will find a solution. There are two ways of uploading your own data to MorphoNet: isolated time points can be uploaded with a dedicated Fiji plugin: https://morphonet.org/help_fiji.
More complex datasets (e.g. time series) can be uploaded through the python API: https://morphonet.org/help_api
What type of data can I upload and visualize?
Although users can upload and visualize raw intensity images, MorphoNet’s full potential is unleashed when working with 3D (+ time) segmented data. If you have not yet generated meshes for your segmentations, the system will automatically perform this task for you.
MorphoNet actually offers more flexibility than genome browsers. While the DNA base pair is the universal unit of information in genome browsers, the choice of the relevant morphological unit of information (i.e., the scale of the segmented objects) in MorphoNet is left to the user, from whole organs down to molecular complexes (Figure 1). Like any web-based system MorphoNet has some limitations, which are progressively lifted as the webGL technology is progressing. It is currently difficult to visualize tissues of embryos with more than a couple of thousand segmented cells or objects.
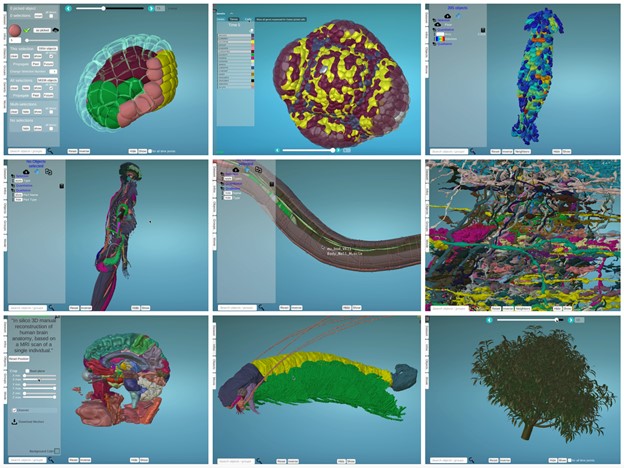
– top left : Astec-Pm1 (Wild type Phallusia mammillata embryo, live SPIM imaging, stages 8-17)
– top middle: Floral Meristem Atlas, stage 0 – 4
– top right: Nest of termites Cubitermes (X-ray tomography)
-middle left: Human body structure (Healthy adult, full-body MRI)
-middle middle: Caenorhabditis elegans (Wild type, adult)
-middle right: Drosophila melanogaster medulla connectome (Wild type, TEM imaging, adult)
-bottom left : Atlas of human brain (Healthy adult, MRI)
-bottom middle: Cascolus ravitis (Fossil, digitally captured images)
-bottom right: Mango tree (Simulated, growth)
Can I see inside the imaged object?
Uploaded datasets are fully interactive. You can rotate, zoom in or out, and click on each individual object. To explore the inside of your segmented structures, you can easily remove entire cell layers, or individually select and hide individual objects. You can also use the scatter view to literally explode all objects radially to clearly visualize each of them (See below).
Can I simultaneously visualise different types of segmented objects (e.g., nuclei within whole cells)?
Yes. You can upload datasets with several segmented channels, obtained from membrane and nuclei recordings, for example. You can also directly add your raw intensity images to your scene. This can be very useful to see gene expression over cell segmentation for example.
Can I annotate my dataset with MorphoNet?
Yes. MorphoNet can be used to add different types of annotations to your dataset: temporal (i.e. to link objects through time), qualitative (for example, to name objects), quantitative (to attribute a value to each individual element, for example cell volume (see below)), a selection value (a given number for each objects) and others (details of the formats used can be found here https://morphonet.org/help_format ). You can either directly label your objects in the scene or upload an annotation file.
Can I assess the quality of my segmentation with the original microscope images?
Yes, you can upload the original microscope images used to segment your dataset and compare them with your segmented data. You can either display the full 3D volume using a threshold value to remove the background or just show some slices in X, Y or Z. You can have a look at this example.
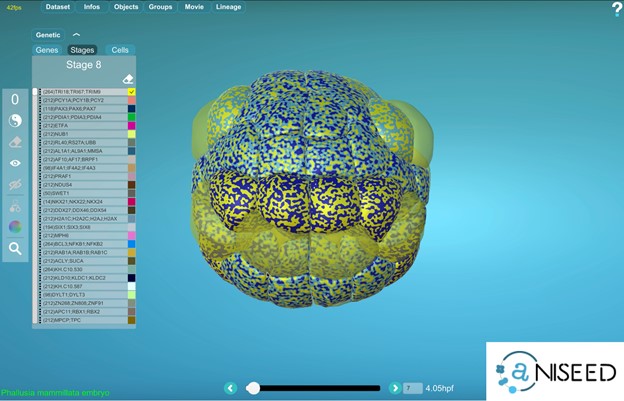
Can I use MorphoNet to visualize Gene Expression Data?
Yes, for ascidian embryos, MorphoNet is interfaced to the gene expression section of the ANISEED database (https://www.aniseed.cnrs.fr) (See Figure 2 and movie below). Other external gene expression databases could be connected to MorphoNet in the future, and we will be happy to work with you on such projects. If no gene expression database is available for your organism, you can directly upload your expression profiles using a simple format (https://morphonet.org/help_format) and then, as we did with the Arabidopsis meristem (3), you can manually curate, if necessary, your gene expression information by labeling individual cells.
To explore gene expression data, each individual gene can be assigned a colour, and the powerful MorphoNet shaders allow you to simultaneously visualize the expression profiles of up to 4 genes. You can also interrogate your data for genes expressed in the intersection or the union of some specific selected cells.
Can I explore the cell lineage?
MorphoNet is a unique tool which allows you to interact at the same time with your 3D+t dataset and the corresponding lineage. We use a second window to display the cell lineage and each click on a specific cell will also be displayed on the lineage. You can also map supplementary information, such as the cell volume, on the lineage and then order the branches by these values.
How can I make nice figures from MorphoNet?
We recently implemented a new module (inside the tools menu), which allows you to create your own figures in pdf format. You can also create your own movies in MorphoNet. Simply define some key frames and MorphoNet will interpolate the rest.
How can I share data with my colleagues?
You can easily share a specific dataset you own with any colleague who registered to MorphoNet. If you want to share multiple datasets, you can group them into a working group, which can be shared. You can also use MorphoNet to share data publicly, in an article for instance. For this, you can create a session, with a permanent url that can be shared (Just click here as an example https://morphonet.org/hgCbNDIV).
Can I do more if I can code?
You can directly query your data using the python API : https://morphonet.org/help_api. It is very useful when you have many datasets or when you automatically compute some properties. We are also developing a Plot module which allows us to directly visualize your data inside our 3D viewer without any data upload. And you can, of course, contribute! MorphoNet is an open-source software. Please contact us if you are interested. We are having a lot of fun.
What are the next features that you plan to add in MorphoNet?
We have several projects underway. We are currently working on a standalone application which you can directly install on your computer to fully exploit its computational resources when interacting with big datasets.
We are also working on the Virtual Reality representation of your data. This will allow you to interact with your own data with a classical VR headset.
Stay tuned! You can follow us on Twitter @MorphoTweet
REFERENCES:
- B. Leggio, J. Laussu, A. Carlier, C. Godin, P. Lemaire, E. Faure, MorphoNet : an interactive online morphological browser to explore complex multi-scale data. Nat Commun. 10, 1–8 (2019).
- L. Guignard, U.-M. Fiúza, B. Leggio, J. Laussu, E. Faure, G. Michelin, K. Biasuz, L. Hufnagel, G. Malandain, C. Godin, P. Lemaire, Contact area–dependent cell communication and the morphological invariance of ascidian embryogenesis. Science. 369, aar5663 (2020).
- Refahi et al. A multiscale analysis of early flower development in Arabidopsis provides an integrated view of molecular regulation and growth control. Developmental Cell, Elsevier, 2021, 56 (4), pp.540-556.e8. ⟨10.1016/j.devcel.2021.01.019⟩


 (No Ratings Yet)
(No Ratings Yet)