Featured Resource: The Arabidopsis Information Resource
Posted by Tanya Berardini, on 12 October 2022
Doing great science depends on teamwork, whether this is within the lab or in collaboration with other labs. However, sometimes the resources that support our work can be overlooked. Our ‘Featured resource’ series aims to shine a light on these unsung heroes of the science world. In our latest article, we hear from Tanya Z. Berardini, Ph.D (TAIR Director) and Leonore Reiser, Ph.D (Senior Scientific Curator)] who describe the work of TAIR.
The Arabidopsis Information Resource (TAIR) was established in 1999 with US National Science Foundation funding and the goal of creating a database of genetic and molecular biology data for the model higher plant Arabidopsis thaliana. Arabidopsis was the first plant genome to be sequenced (1). Since then, over twenty years of research have continually improved the sequence and functional annotation of a genome that serves as a reference for an ever-increasing number of newly sequenced plant genomes (2). TAIR integrates, interconnects, and consolidates information from peer-reviewed published literature with sequence and stock information (3) so that researchers can spend less time searching for information and more time developing and testing hypotheses guided by work that has already been done.
Who runs the resource?
TAIR was created at the Department of Plant Biology of the Carnegie Institute for Science and the National Center for Genome Resources (NCGR) and supported by the NSF from 1999 to 2013. Since 2014, it has been administered and maintained by the non-profit organization, Phoenix Bioinformatics (4). Phoenix has a small team of dedicated data curation scientists, who meticulously curate and update TAIR data, and software engineers, who maintain and update TAIR’s database and tools. Newly curated information from the literature is added weekly.
Where does funding come from?
Since 2014, TAIR’s operations have been funded by subscriptions from academic and other non-profit institutions, individual researchers, corporations and countries (e.g., China through its National Science and Technology Library). Institutional subscriptions are priced based on past year usage, so that institutions that use the resource more contribute more to its maintenance and growth. TAIR’s subscription support is as global as its user base. Phoenix Bioinformatics has additional funding from the NSF and the Sloan Foundation for other aspects of its work.
Can one access TAIR without a subscription?
Unlimited access to TAIR’s data and tools almost always requires a subscription. However, TAIR provides a limited number of free page views for occasional users each month. This is similar to the monthly allotment of complimentary articles often offered by online newspapers and magazines. After the monthly limit is reached a subscription is encouraged. Upon request, TAIR grants full access for teachers and students at non-subscribing institutions for classes that use TAIR in their curriculum. US Historically Black Colleges and Universities (HBCUs) are granted free access. Finally, we grant country-wide access to those countries that fall into the ‘low-income economies’ categorization of the World Bank.
What tools and resources are available for researchers?
- The TAIR locus page is a treasure trove that consolidates in-house data, data pulled from other resources by APIs, and links to external resources with complementary information. New articles, gene symbols and full names, gene summaries, Gene Ontology (GO) and Plant Ontology (PO) annotations, germplasms and phenotypes based on those articles are added by data curation scientists to a subset of loci on a weekly basis.
- Researchers can find information for single genes or they can download data (sequences, descriptions, GO annotations) for sets of genes or even the whole genome.
- There are two types of quarterly data releases: (A) Public data releases are a year old and are released for public reuse with a CC-BY reuse license. (B) Subscriber data releases reflect the most recent year’s data.
- TAIR provides data search and browsing services, data analysis tools, and data visualization tools, such as genome browsers (e.g., JBrowse) and a BLAST service that includes unique datasets.

How can the community contribute?
- Make data FAIR (5).
- Use AGI identifiers for the Arabidopsis genes in papers!
- Use stock identifiers from NASC and ABRC for seed and DNA stocks. They allow us to link these stocks unambiguously to locus records at TAIR.
- Register gene symbols, use the symbols for genes that already exist and check that symbols you want to use are not already in use.
- Submit functional annotation data using GOAT (goat.phoenixbioinformatics.org).
- Contribute genomic coordinate-anchored datasets that can be visualised in JBrowse.
- Cite TAIR if you have used it in your work. Guidelines are available here.
- Email us at curator@arabidopsis.org or make comments on pages to report errors and/or omissions.
- Respond to requests for information from TAIR.
- Provide feedback on our in-progress beta.arabidopsis.org site by email.
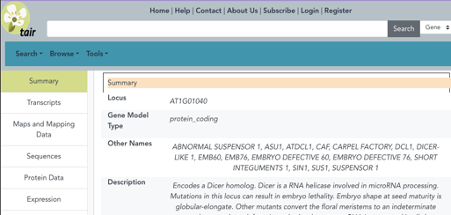
Any hidden gems, features that are new, or that researchers might be less aware of?
- The biggest hidden gem is that if you email us, we will respond within 24 hrs during a regularly scheduled work week.
- Not only can one view information for one gene at a time, one can also upload a list of genes and retrieve gene descriptions, sequences, and GO annotations (and other types of data) in bulk.
- The TAIR YouTube channel has tutorials and webinars of different lengths explaining various features of the resource. Watch a video and learn something new.
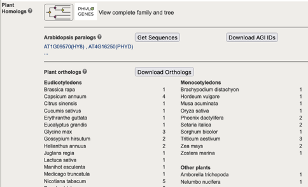
- Access to PhyloGenes (www.phylogenes.org) is integrated into the TAIR Locus page. You can link to the Panther-based gene family, explore experimental and phylogenetically-inferred functional information for gene family members in Arabidopsis and 29 other plants, and take advantage of experimental annotations made in Arabidopsis and other model organisms.
- The TAIR Job listings page is very active with about three new listings posted a week. Some labs recruit almost exclusively by posting their grad student and post-doc openings at https://www.arabidopsis.org/news/jobs.jsp. All openings are also shared through our Twitter account (@tair_news).
References:
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 408, 796–815 (2000). doi.org/10.1038/35048692
- Nicholas J Provart, Siobhan M Brady, Geraint Parry, Robert J Schmitz, Christine Queitsch, Dario Bonetta, Jamie Waese, Korbinian Schneeberger, Ann E Loraine. Anno genominis XX: 20 years of Arabidopsis genomics, The Plant Cell. 33(4):832–845 (2021). doi.org/10.1093/plcell/koaa038
- Tanya Z. Berardini, Leonore Reiser, Donghui Li, Yarik Mezheritsky, Robert Muller, Emily Strait, Eva Huala. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis. 53(8):474-85 (2015). doi.org/10.1002/dvg.22877
- Leonore Reiser, Tanya Z. Berardini, Donghui Li, Robert Muller, Emily M. Strait, Qian Li, Yarik Mezheritsky, Andrey Vetushko, Eva Huala. Sustainable funding for biocuration: The Arabidopsis Information Resource (TAIR) as a case study of a subscription-based funding model, Database. Volume 2016, (2016). doi.org/10.1093/database/baw018
- Leonore Reiser, Lisa Harper, Michael Freeling, Bin Han, Sheng Luan. FAIR: A Call to Make Published Data More Findable, Accessible, Interoperable, and Reusable, Molecular Plant 11(9):1105-1108 (2018). doi.org/10.1016/j.molp.2018.07.005
Contributed by Tanya Z. Berardini, Ph.D (TAIR Director) and Leonore Reiser, Ph.D (Senior Scientific Curator)


 (No Ratings Yet)
(No Ratings Yet)