Featured Resource: the Zebrafish Information Network (ZFIN)
Posted by the Node, on 17 December 2021
Doing great science depends on teamwork, whether this is within the lab or in collaboration with other labs. However, sometimes the resources that support our work can be overlooked. In our new series, ‘Featured Resource’, we aim to shine a light on these unsung heroes of the science world. For our third article, Yvonne Bradford and Leyla Ruzicka tell us about the work of ZFIN. Read on to find out a little about the history of ZFIN, the people running the site and what resources are available.
When was ZFIN established and how has it evolved?
The Zebrafish Information Network (ZFIN) was established by Monte Westerfield in 1994 following the first international Zebrafish research meeting at Cold Spring Harbor. In its infancy, ZFIN played a large role as a community contact resource for zebrafish researchers and students. By 1997 ZFIN curators were establishing nomenclature guidelines for genes and mutant lines, as well as curating mutant line information from scientific publications1. Leap to 2003 and ZFIN had established cross links and integration of ZFIN data with other genomic resources including GenBank, Vega, UniProt and Ensembl, and curators were annotating additional data types that included gene expression, orthology, and Gene Ontology data2. Over the next several decades, ZFIN curators continued to expand the data supported at ZFIN to include complex genotypes, transgenic lines, phenotypes, morpholinos, CRISPRs, TALENs, antibodies, and transcripts34567. Over the past 27 years, ZFIN has grown into a mature knowledgebase for zebrafish researchers providing information about genes and alleles, gene expression and phenotype data, human disease associations and models, Gene Ontology data for gene function, orthology, reagent information and protocol wikis, BLAST and Genome Viewers, as well as deep integration with genetic and genomic data service providers. Throughout ZFIN’s history, serving the research community has been central to the development of the resource. ZFIN has a core aim of providing accurate, accessible and interoperable data to researchers, students, bioinformaticians, and other online databases to facilitate the use of Danio rerio as a model organism to understand developmental biology, genetics, genomics, phenomics, and ultimately human biology.
Who are the people behind the resource?
ZFIN is located at the University of Oregon in Eugene, Oregon. The ZFIN team consists of scientific curators who hold a PhD or MSc and have expertise in genetics, developmental, cellular, molecular, and evolutionary biology, as well as software developers skilled in Java web architecture, PostgreSQL database development, and many modern coding languages to implement user interfaces. Although the following list represents ZFIN staff as of 2021, many prior staff members significantly contributed to the success of ZFIN over the years.
- Principal Investigator – Monte Westerfield
- Project Manager – Yvonne Bradford
- Scientific Curation Team:
- David Fashena
- Ken Frazer
- Doug Howe
- Holly Paddock
- Sridhar Ramachandran
- Leyla Ruzicka
- Amy Singer
- Wei-Chia Tseng
- Ceri Van Slyke
- Software Development Team:
- Ryan Martin
- Christian Pich
- Ryan Taylor
- Literature Acquisition Assistant – Kaia Fullmer
Where does funding come from?
Initially, ZFIN was funded by the W.M Keck Foundation and the National Science Foundation. In 1998, Dr. Westerfield was awarded NIH funding for the Zebrafish International Resource Center (ZIRC) that included database support for ZFIN. Since 2002, ZFIN has been independently funded by the National Human Genome Research Institute of the National Institutes of Health.
What is available for researchers?
ZFIN hosts a diverse collection of data, as well as genomic and community resources. ZFIN provides expertly curated data on zebrafish, including information on genes, alleles, transgenic lines, human disease models, gene expression, mutant phenotypes, gene function, orthology, publications, researchers, labs, and reagents. ZFIN has data-specific web pages for genes, alleles, fish, antibodies, constructs, morpholinos, CRISPRs, TALENs, clones, publications, and ontology terms. Researchers can search for data using the single box search on the home page (https://zfin.org/) with results returned in the single box search interface, which allows for easy refinement of results using filters. ZFIN data are also available for download (https://zfin.org/downloads). ZFIN is the nomenclature authority for zebrafish genes, alleles, and transgenic lines. ZFIN provides nomenclature support to the community through the ZFIN Nomenclature Coordinator who works with the Zebrafish Nomenclature Committee, which is composed of active zebrafish researchers, to approve names and symbols for gene loci and transgenic lines. Researchers can contact the ZFIN Nomenclature Coordinator at nomenclature@zfin.org.
ZFIN provides access to genomic resources through the Genomics menu on the Home Page. Researchers can access BLAST resources and genome browsers at ZFIN, NCBI, UCSC, and Ensembl. In addition, the ZFIN gene page has genome browser images and links to sequence analysis tools in the sequences and transcripts sections.
Researchers can access community resources from the Resources and Community menus at the top of the ZFIN Home Page. The Resources menu provides links to The Zebrafish Book, Anatomy Atlases, Zebrafish Resource Centers, and community wikis. ZFIN hosts the zebrafish research community wiki that has sections for protocols and antibodies. The protocols section provides an area where zebrafish researchers can share experimental protocols and tips covering a range of topics from general zebrafish care to molecular and behavioural methods, and the antibodies section focuses on antibody usage notes and protocols. The Community menu offers researchers access to zebrafish News, Job postings, and Meeting announcements, as well as links to search for researchers, labs, and companies.
How can the community contribute to ZFIN?
In addition to annotating zebrafish research publications, ZFIN accepts direct submissions from users. From the Support menu at the top of the home page, under ‘Using ZFIN’, there is a link to submit data to ZFIN. The link leads to a page that has links to the various ways users can submit data directly. To help researchers submit data, a guide that outlines the minimal set of information for data that are submitted for inclusion in ZFIN is available in the Methods in Cell Biology chapter: A Scientist’s guide for submitting data to ZFIN8.
How can the community help ZFIN?
The research community can help ZFIN in many ways:
- Cite ZFIN in publications. Directions for citing ZFIN can be found in the Support menu, in the About ZFIN link (https://zfin.atlassian.net/wiki/spaces/general/pages/1891415775/ZFIN+Database+Information)
- Respond to ZFIN surveys. ZFIN sends out user surveys to registered users yearly. The surveys determine which features will be developed at ZFIN
- Keep your person/lab/company pages in ZFIN up to date
- Help facilitate the curation of your publications:
- Contact ZFIN about gene and allele/transgenic line nomenclature before publication
- Make sure data in papers are unambiguous – use identifiers, accession numbers, and approved names whenever possible for genes, alleles, and reagents.
- Respond to requests for information by ZFIN curators on details in papers including genes, alleles, transgenes, sequence information, and reagents.
What are the most used features on ZFIN?
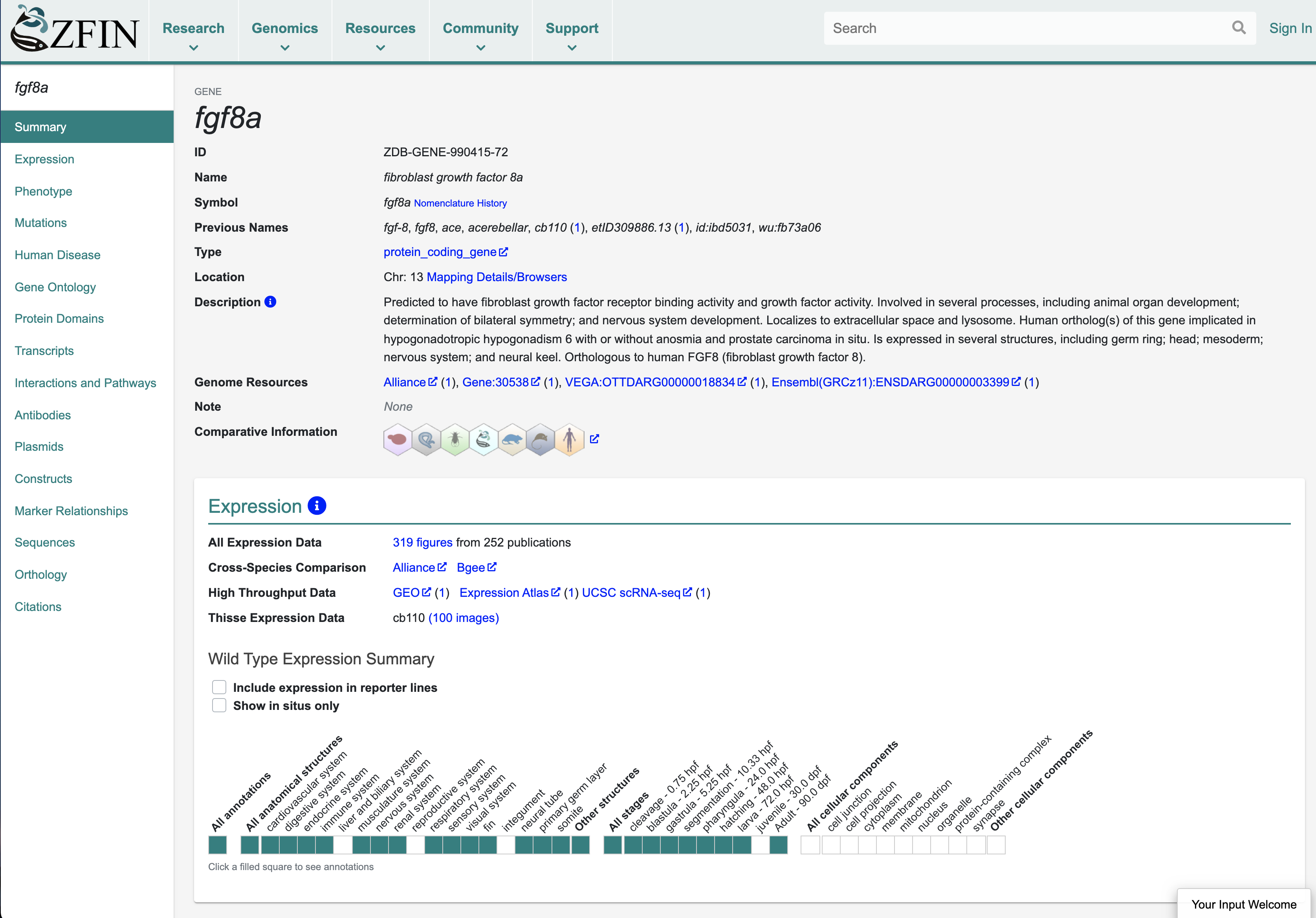
Gene pages are the most highly accessed pages at ZFIN. The gene page integrates all data pertinent to a gene and is a hub of information for researchers. Other ZFIN resources that are highly accessed include gene expression and phenotype data, publications and associated images, and search interfaces. ZFIN person and lab pages are also very popular, as is The Zebrafish Book9, the classic handbook for zebrafish research.
Any new developments that you can recommend, which researchers might be less aware of?
ZFIN is a founding member of the Alliance of Genome Resources (The Alliance), a consortium of six Model Organism databases (Mouse Genome Database, Rat Genome Database, Saccharomyces Genome Database, WormBase, FlyBase, and ZFIN) and the Gene Ontology database. The Alliance aims to develop and maintain sustainable genome information resources that facilitate the use of model organisms in understanding the genetic and genomic basis of human biology and disease10. Follow the “Alliance” links from ZFIN gene pages to explore comparative data on Orthology, Human Disease, Gene Ontology, and Gene Expression, plus details about Variants and Phenotypes. The Alliance also provides download files and APIs for programmatic access to model organism data.
References
1. Westerfield M, Doerry E, Kirkpatrick A, Driever W, Douglas S. An on-line database for zebrafish development and genetics research. Seminars in cell & developmental biology. 1997;8(5):477–88. doi:10.1006/scdb.1997.0173
2. Sprague J, Clements D, Conlin T, Edwards P, Frazer K, Schaper K, Segerdell E, Song P, Sprunger B, Westerfield M. The Zebrafish Information Network (ZFIN): the zebrafish model organism database. Nucleic acids research. 2003;31(1):241–243. doi:10.1093/NAR/GKG027
3. Sprague J, Bayraktaroglu L, Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Haendel M, Howe DGDG, Knight J, et al. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic acids research. 2008;36(Database issue):D768-72. doi:10.1093/nar/gkm956
4. Bradford YM, Conlin T, Dunn NA, Fashena D, Frazer K, Howe DG, Knight J, Mani P, Martin R, Moxon SAT, et al. {ZFIN:} enhancements and updates to the zebrafish model organism database. Nucleic Acids Res. 2011;39(Database-Issue):822–829. doi:10.1093/nar/gkq1077
5. Howe DG, Bradford YM, Conlin T, Eagle AE, Fashena D, Frazer K, Knight J, Mani P, Martin R, Moxon SAT, et al. ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics. Nucleic Acids Research . 2013;41(Database issue):D854-60. doi:10.1093/nar/gks938
6. Ruzicka L, Bradford YM, Frazer K, Howe DG, Paddock H, Ramachandran S, Singer A, Toro S, Van Slyke CE, Eagle AE, et al. ZFIN, The zebrafish model organism database: Updates and new directions. Genesis. 2015;53(8). doi:10.1002/dvg.22868
7. Howe DG, Ramachandran S, Bradford YM, Fashena D, Toro S, Eagle A, Frazer K, Kalita P, Mani P, Martin R, et al. The zebrafish information network: Major gene page and home page updates. Nucleic Acids Research. 2021;49(D1):D1058–D1064. doi:10.1093/nar/gkaa1010
8. Howe DGG, Bradford YMM, Eagle A, Fashena D, Frazer K, Kalita P, Mani P, Martin R, Moxon STT, Paddock H, et al. A scientist’s guide for submitting data to ZFIN. Methods in Cell Biology. 2016;135:451–481. doi:10.1016/bs.mcb.2016.04.010
9. Westerfield M. The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio). 4th ed. Eugene, OR: University of Oregon Press; 2000.
10. Bult CJ, Blake JA, Calvi BR, Cherry JM, DiFrancesco V, Fullem R, Howe KL, Kaufman T, Mungall C, Perrimon N, et al. The alliance of genome resources: Building a modern data ecosystem for model organism databases. Genetics. 2019;213(4):1189–1196. doi:10.1534/genetics.119.302523


 (No Ratings Yet)
(No Ratings Yet)