Flippase recognition target: orientation matters, so why care?
Posted by Mario Metzler, on 16 April 2014
FRT sites are used often (at least in Drosophila) for inducing deletions or “flipping out” of markers in transgenic constructs.
When there are two FRTs sequences in tandem, after inducing flippase the DNA sequence that is between these two sites will be deleted. If two FRT sites are facing each other (or looking away), the DNA that sits in between them can be inverted after induction of flippase.
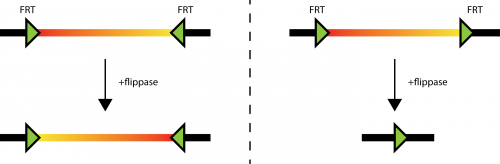 Fig. 1: different behavior of flippase induced recombination of FRT sites. Left: sin the situation where two FRT sites are in different orientation, the DNA laying between them (the thick line with a red to yellow coloration) can be inverted. As the FRT sites are not deleted in the process, the inversion can happen many times. Right: If the FRT sites sit in tandem on the DNA (thick line), the DNA laying in between them can be deleted. Only one FRT site is left on the original DNA.
Fig. 1: different behavior of flippase induced recombination of FRT sites. Left: sin the situation where two FRT sites are in different orientation, the DNA laying between them (the thick line with a red to yellow coloration) can be inverted. As the FRT sites are not deleted in the process, the inversion can happen many times. Right: If the FRT sites sit in tandem on the DNA (thick line), the DNA laying in between them can be deleted. Only one FRT site is left on the original DNA.
Figure 1 illustrates how important the orientation of the FRT sites is for experimental design. So there should be a convention on how to label the orientation of those elements (as I did in the figure above, where the arrow shows from 5′ end to 3′ end of the element). Most scientist draw the FRT sites in a similar way on their plasmid maps. The big problem is, that this is not done in a consistent way.
The wikipedia article on FRT states the following:
5′GAAGTTCCTATTCtctagaaaGtATAGGAACTTC3′
This is a clear definition of orientation, and the article writers took this information out of a paper published in 1994 (Schlake and Bode, Biochemistry 33 (43): 12746–12751).
Others use a different annotation of the FRT element, where exactly the other strand is the leading strand (for example in “Drosophila, a laboratory handbook”, Ashburner et al., second edition).
This can cause some problems, especially when sharing plasmids and flies between laboratories or even between people in the same lab. One has to be consistent, and not trust the graphic map of a plasmid, but its sequence.
I hope this blog post helps other scientists to prevent the bad luck I had with experimental design. Always check the sequence and be consistent :)


 (11 votes)
(11 votes)
This blog was short but precise,it is very useful for me,thank you very much!