In Development this week (Vol. 138, Issue 19)
Posted by Seema Grewal, on 6 September 2011
Here are the research highlights form the current issue of Development:
Modelling liver development with ES cells: HNF4A is key

Human embryonic stem cells (hESCs), via their ability to differentiate into a plethora of cell types, offer an attractive approach for regenerative medicine, but they also offer a means of studying cell differentiation, and hence development, ex vivo. Here, Stephen Duncan and co-workers analyse the differentiation of hESCs to probe the molecular mechanisms that underlie human hepatocyte differentiation (see p. 4143). Using a protocol in which hESCs differentiate into hepatocytes in a stepwise manner, the researchers show that each stage of the differentiation process is associated with a characteristic mRNA profile, as shown by microarrays. Importantly, they show that the transcription factor HNF4A, which has been implicated in liver development, is essential for specifying hepatic progenitors; the onset of HNF4A expression is associated with specification of the hepatic lineage from hESCs, and shRNA-mediated knockdown of HNF4A prevents hESC differentiation into hepatic progenitors. These and other studies demonstrate that HNF4A establishes the expression of a network of transcription factors that promote hepatocyte cell fate.
Gcm/Glide-ing to a glial fate
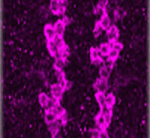
Neurons and glia originate from a common precursor, the neural stem cell (NSC). These multipotent precursors display a high degree of plasticity in vitro, but the basis of this plasticity and the mechanisms underlying the neuron-glial switch in vivo are unclear. Now, Angela Giangrande and colleagues (see p. 4167) show that the transcription factor Glial cells missing (Gcm, also called Glial cell deficient; Glide) triggers a conserved chromatin signature that converts Drosophila NSCs to a glial fate. The researchers show that overexpression of Gcm in fly NSCs produces glia at the expense of neurons. This gliogenic potential of Gcm decreases with time and does not affect quiescent NSCs, suggesting that it is dependent on temporal cues rather than on the mitotic potential of NSCs. Finally, the investigators demonstrate that the glial fate switch is associated with a chromatin signature, which includes low levels of histone H3 lysine 9 acetylation and is similar to that observed in vertebrate glia, suggesting that this epigenetic mechanism for specifying glia has been conserved throughout evolution.
Paired up: histone methylation and HP1γ
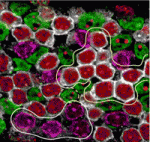
The pairing of chromosomes during meiosis requires histone modifications, such as histone H3 lysine 9 di- and tri-methylation (H3K9me2 and H3K9me3, respectively), at pericentric heterochromatin (PCH) regions. But how do these epigenetic marks control chromosome interactions? Haruhiko Koseki and colleagues demonstrate that heterochromatin protein 1γ (HP1γ) regulates chromosome interactions by recognising histone methylation marks during meiosis in mice (see p. 4207). The researchers show that, in meiotic spermatocytes, H3K9me2 by the G9a histone methyltransferase requires pre-existing H3K9me3 marks, which are deposited by the Suv39h histone methyltransferase. They further show that HP1γ recognizes H3K9me3 marks and localises to PCH regions in an H3K9me3-dependent manner, where it then recruits G9a. Importantly, the loss of HP1γ results in defective spermatogenesis, aberrant centromere clustering and impaired homologous chromosome pairing. The authors thus propose that HP1γ acts as an important link between the cascade of H3K9me3 and H3K9me2 modifications, acting to align homologous chromosomes and facilitate their pairing during meiosis.
A new look into Sfrps and Wnt
 Secreted frizzled-related proteins (Sfrps) are classified as Wnt antagonists, but recent studies have shown that some Sfrps can positively modulate Wnt signalling. Is this a general property of all Sfrps and, if so, how do Sfrps regulate Wnt signalling? Here, Paola Bovolenta and colleagues (see p. 4179) show that Sfrp1 and Sfrp2 positively regulate Wnt signalling, and are required for Wnt-mediated development of the mouse optic cup. The researchers show that specification of the peripheral optic cup (OCP), which is known to be dependent on Wnt signalling, is grossly defective in mice lacking both Sfrp1 and Sfrp2. In these mutants, Wnt spreading across the OCP is impaired, suggesting that Sfrps can influence the diffusion of Wnts. In support of this, the researchers demonstrate that Sfrp1 overexpression flattens the gradient of Wingless (a Drosophila Wnt homologue) across the Drosophila imaginal disc. These studies highlight a new and unexpected role for Sfrps in regulating the levels and distribution of Wnts during development.
Secreted frizzled-related proteins (Sfrps) are classified as Wnt antagonists, but recent studies have shown that some Sfrps can positively modulate Wnt signalling. Is this a general property of all Sfrps and, if so, how do Sfrps regulate Wnt signalling? Here, Paola Bovolenta and colleagues (see p. 4179) show that Sfrp1 and Sfrp2 positively regulate Wnt signalling, and are required for Wnt-mediated development of the mouse optic cup. The researchers show that specification of the peripheral optic cup (OCP), which is known to be dependent on Wnt signalling, is grossly defective in mice lacking both Sfrp1 and Sfrp2. In these mutants, Wnt spreading across the OCP is impaired, suggesting that Sfrps can influence the diffusion of Wnts. In support of this, the researchers demonstrate that Sfrp1 overexpression flattens the gradient of Wingless (a Drosophila Wnt homologue) across the Drosophila imaginal disc. These studies highlight a new and unexpected role for Sfrps in regulating the levels and distribution of Wnts during development.
Kidney development: a Notch above the Wnts
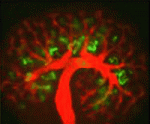 The kidney comprises functional units known as nephrons, which are made up of specialised epithelial cells. During development, each nephron arises from a pool of stem cells that undergo mesenchymal-to-epithelial transition (MET) in response to signals such as Wnt4 and Wnt9b. Here, Raphael Kopan and co-workers show that Notch pathway activation can replace inductive Wnt signals during this process (see p. 4245). Using gene manipulation in cultured kidneys, the researchers show that Notch pathway activation can induce epithelialisation in nephron stem cells but not in the closely related stromal mesenchymal cells. Continued Notch pathway activation following MET directs cells towards a proximal tubule fate. Finally, they report, Notch-induced MET can occur in the absence of Wnt4 and Wnt9b, suggesting that nephron stem cells are poised to undergo MET, which requires a permissive signal that can be provided by Wnts or by Notch pathway activation. These studies shed new light on our understanding of the early cell fate decisions that are made during kidney development.
The kidney comprises functional units known as nephrons, which are made up of specialised epithelial cells. During development, each nephron arises from a pool of stem cells that undergo mesenchymal-to-epithelial transition (MET) in response to signals such as Wnt4 and Wnt9b. Here, Raphael Kopan and co-workers show that Notch pathway activation can replace inductive Wnt signals during this process (see p. 4245). Using gene manipulation in cultured kidneys, the researchers show that Notch pathway activation can induce epithelialisation in nephron stem cells but not in the closely related stromal mesenchymal cells. Continued Notch pathway activation following MET directs cells towards a proximal tubule fate. Finally, they report, Notch-induced MET can occur in the absence of Wnt4 and Wnt9b, suggesting that nephron stem cells are poised to undergo MET, which requires a permissive signal that can be provided by Wnts or by Notch pathway activation. These studies shed new light on our understanding of the early cell fate decisions that are made during kidney development.
Ngn2 phosphorylation links neurogenesis to the cell cycle
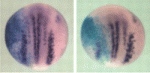
Cell cycle length influences the balance between progenitor maintenance and differentiation in the nervous system, although the mechanism for this is unknown. Here, Anna Philpott and co-workers show that multi-site phosphorylation of neurogenin 2 (Ngn2), a master regulator of neuronal development, controls neuronal differentiation in response to cell cycle lengthening in Xenopus embryos and in mammalian P19 cells (see p. 4267). The researchers show that, in Xenopus extracts, Ngn2 phosphorylation is regulated by the cell cycle, and analyses of HeLa cell extracts show that Ngn2 is phosphorylated on multiple sites by cyclin-dependent kinases (cdks). The phosphorylation of Ngn2, they report, reduces its ability to induce neuronal differentiation in vivo, and this is due to the decreased ability of phosphorylated Ngn2 to bind to its target promoters. The authors thus propose a model in which multi-site phosphorylation of Ngn2, which is quantitatively sensitive to cell cycle length, is used as a way to interpret cdk levels in order to control neuronal differentiation in response to cell cycle lengthening during development.
Plus…
The control of developmental phase transitions in plants
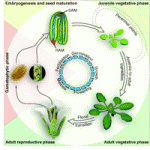
Plant development progresses through distinct phases, each controlled by genetic pathways that integrate endogenous and environmental cues. Recent studies, reviewed by Huijser and Schmid, show that the genetic networks underlying the transitions between these phases share some common factors.
See the Review article on p. 4117
Modeling new conceptual interpretations of development
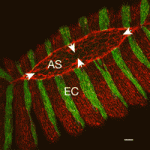
As reviewed by Julien Vermot and Markus Affolter, the EMBO workshop on Biophysical Mechanisms of Development (organized by Ana Borges, Ana Certal, Ana Tavares, Filipa Alves and Beatriz Garcia Fernandez) brought together scientists in the field of developmental biology for whom interdisciplinary and quantitative approaches are central to the issues they are investigating.
See the Meeting Review, p. 4111


 (1 votes)
(1 votes)