In Development this week (Vol. 138, Issue 5)
Posted by Seema Grewal, on 8 February 2011
Here are the highlights from the current issue of Development:
From pluripotent to pancreatic fates
A reliable method for generating insulin-producing β-cells from human pluripotent stem cells (hPSCs) would provide new therapeutic options for people with diabetes. So far, no-one has developed such a method but, on p. 861, Gordon Keller and colleagues provide new insights into the complex signalling networks that underlie β-cell differentiation. The generation of β-cells from hPSCs requires efficient endoderm induction followed by patterning and specification to a pancreatic fate. The researchers show that the duration of nodal/activin A signalling plays a pivotal role in endoderm induction and that WNT signalling enhances the subsequent development of pancreatic lineage cells. Inhibition of BMP signalling at specific stages is also essential for the generation of insulin-expressing cells. Importantly, report the researchers, optimal stage-specific manipulation of TGFβ and WNT signalling yields cell populations that produce insulin at levels similar to those made by the pancreas. However, because these cells also make other hormones, further studies are needed to discover how to convert these polyhormonal cells into functional β-cells.
Full Fat signalling in mammals: Dchs1 and Fat4 pair up

In Drosophila, Dachsous and Fat act as ligand and receptor, respectively, for a signalling pathway that regulates planar cell polarity (PCP) and transcription via the Hippo pathway. Mammals encode multiple Fat and Dachsous proteins but do they have an equivalent Fat signalling pathway? On p. 947, Yaopan Mao and colleagues report that murine Dchs1 and Fat4 function as a ligand-receptor pair during development. The researchers show that Dchs1 and Fat4 single mutants and Dchs1 Fat4 double mutants exhibit similar phenotypes. These phenotypes include the formation of kidney cysts and cochlear defects, suggesting that Dchs1-Fat4 signalling influences PCP in mice. However, the researchers also identify non-PCP-related requirements for Dchs1-Fat4 signalling in the development of other organs. In particular, they show that Dchs1 and Fat4 are needed for growth, branching and cell survival during early kidney development. Together, these results identify Dchs1 and Fat4 as a ligand-receptor pair for mammalian Fat signalling and identify new requirements for Fat signalling in multiple organs.
Capicua mediates response to RTK signalling
Receptor tyrosine kinase (RTK) signalling pathways regulate many developmental decisions, but how RTK signalling controls the expression of its target genes in different contexts is poorly understood. Here, Gerardo Jiménez and co-workers reveal that octameric DNA-binding sites for the transcriptional repressor Capicua (Cic) are critically involved in RTK signalling in Drosophila (see p. 915). They show that the regulation of terminal gap gene expression by the Drosophila RTK Torso in early embryos depends on octameric Cic-binding sites in the enhancer region of the gap gene huckebein. Moreover, these Cic-binding motifs are essential for recruitment of the Groucho co-repressor to the huckebein enhancer in vivo. Cic-binding sites also respond to EGFR RTK pathway activation in the embryonic neuroectoderm and in the developing wing. Finally, using synthetic enhancer constructs, the researchers show that Cic-binding motifs provide the regulatory information necessary to translate RTK signalling inputs into precise transcriptional responses in different tissues. Thus, they conclude, octameric Cic-binding motifs are general response elements for RTK signalling in Drosophila.
A new spin(dle) on stem cell division
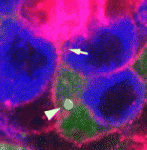
Stem cells divide asymmetrically to balance self-renewal and differentiation, thereby maintaining tissue homeostasis. But what coordinates the divisions of multiple stem cell populations in complex tissues? To address this question, Yukiko Yamashita, Alan Hunt and colleagues (see p. 831) have been studying stem cell division in the Drosophila testis, which contains both germline stem cells (GSCs) and somatic cyst stem cells (CySCs). GSCs divide asymmetrically by maintaining a fixed cell polarity within the stem cell niche. Now, the researchers use time-lapse live imaging to show that CySC asymmetric division involves the repositioning of a randomly located mitotic spindle during or near anaphase onset. Spindle repositioning, they report, requires functional centrosomes, the motor protein Dynein and the actin-membrane linker Moesin, and is required to achieve the high-fidelity asymmetric CySC divisions that maintain both GSC and CySC numbers. The researchers speculate that the use of multiple mitotic schemes may be a general mechanism whereby divisions of different stem cell populations are coordinated in complex tissues.
Eyeing up neuronal circuits
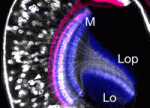
The Drosophila optic lobe shares many characteristics with mammalian visual systems and might provide a powerful model for investigating the formation of visual processing circuits. Little is known, however, about the mechanisms that create neuronal diversity and organise neuronal circuits in the medulla, the optic lobe’s primary region. Now, on p. 983, Makoto Sato and colleagues describe the key features of the developing fly medulla. They show that, during larval development, the medulla is subdivided into concentric zones that are characterised by the expression of the transcription factors Drifter, Runt, Homothorax and Brain-specific homeobox. The birth order of the medulla neurons correlates with the expression pattern of these factors, they report, and each neuronal type exhibits an extensive but defined pattern of migration that disrupts the concentric zones during early pupal development. These results, and those of clonal analyses, lead the researchers to suggest that the concentric zone genes may form a genetic hierarchy that specifies neuronal identity and establishes neuronal circuits in the developing medulla.
Marking up germline imprints
Genomic imprinting – epigenetic modifications that ensure that certain genes are expressed from only one of the two inherited chromosomes – is crucial for normal development. Mammalian imprinted genes are associated with differentially methylated regions (DMRs) that are CpG methylated on one parental chromosome. At least 21 DMRs become methylated in the mouse germline and, on p. 811, Hiroyuki Sasaki and co-workers analyse a panel of these gametic DMRs. The extent of methylation of these DMRs differs significantly from that of embryonic DMRs, they report, suggesting that gametic DMRs should be used to identify the features that establish imprinting in the germline. They also show that maternal gametic DMRs appear as unmethylated islands in male germ cells, and unexpectedly identify widespread oocyte-specific non-CpG methylation. Finally, they report that DMR methylation changes dynamically during early development, indicating that DMRs are not fully protected from preimplantation epigenetic reprogramming. These results underscore the importance of using gametic DMR sequences for the study of imprint establishment.
Also…
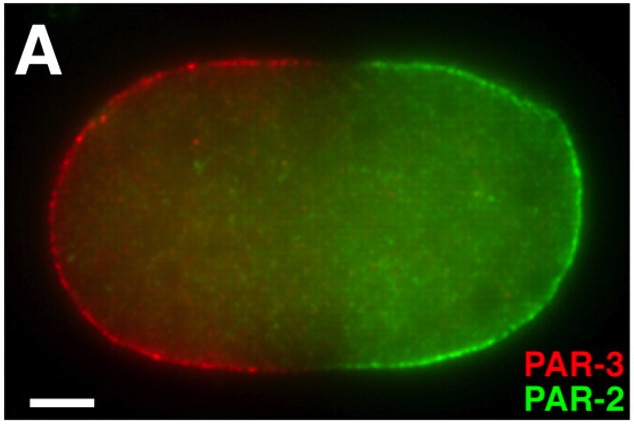
PAR proteins are conserved regulators of cell polarity, and recent studies, reviewed here by Jeremy Nance and Jennifer Zallen, have identified elaborate links between PAR proteins and cytoskeletal proteins that help set-up molecular asymmetries and hence establish polarity within a cell. See the Review on p. 799


 (No Ratings Yet)
(No Ratings Yet)