In Development this week (Vol. 139, Issue 1)
Posted by Seema Grewal, on 6 December 2011
Here are the highlights from the current issue of Development:
Notching up pancreas development
According to the lateral inhibition model, during early pancreas development, Neurog3 expression in multipotent progenitor cells (MPCs) initiates endocrine differentiation and activates expression of the Notch ligand Dll1. Dll1 then activates Notch receptors in neighbouring cells, which turns on Hes1 expression. Finally, Hes1 inhibits Neurog3 expression in these neighbouring cells, thereby preventing excessive endocrine differentiation. On p. 33, Palle Serup and colleagues challenge this model by showing that Dll1, Hes1 and Dll1/Hes1 mutant phenotypes diverge at key points of mouse pancreas development. Moreover, pancreatic hypoplasia in Dll1 mutants is independent of endocrine development and is not, therefore, caused by excessive endocrine differentiation and progenitor depletion, as previously believed. Instead, the researchers report, reduced MPC proliferation is responsible for this hypoplasia. Other results indicate that Ptf1a (pancreas transcription factor 1 subunit α) activates Dll1 expression and that Hes1 sustains Ptf1a expression and Dll1 expression in early MPCs. Thus, Ptf1a-mediated control of Dll1 expression, rather than a lateral inhibition mechanism, is crucial for Notch-mediated control of early pancreas development.
Modelling stem cell population dynamics
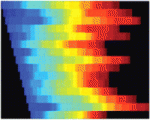
Many tissues and organs are maintained by stem cell populations. Anatomical constraints, cell proliferation dynamics and cell fate specification all affect stem cell behaviour, but their relative effects are difficult to examine in vivo. E. Jane Albert Hubbard, Hillel Kugler and colleagues now use available data to build a computational model of germline development in C. elegans (see p. 47). In this model, germline cells move, divide, respond to signals, progress through mitosis and meiosis, and differentiate according to various local rules. Simulations driven by the model recapitulate C. elegans germline development and the effects of various genetic manipulations. Moreover, the model can be used to make new predictions about stem cell population dynamics. For example, it predicts that early cell cycle defects may later influence maintenance of the progenitor cell population, an unexpected prediction that the researchers validate in vivo. This general modelling approach could, therefore, prove to be a powerful tool for increasing our understanding of stem cell population dynamics.
Time and place generate neuronal diversity
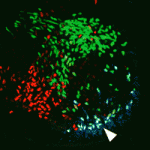
The spinal cord contains many distinct interneuron cell types that have specialised roles in somatosensory perception and motor control. How this neuronal diversity is generated from multipotent progenitor cells is unclear. Here (p. 179), Martyn Goulding and colleagues investigate the differentiation of Renshaw cells, one of the spinal interneuron subtypes that arises from the spatially defined V1 class of interneurons. They show that the first-born postmitotic V1 interneurons are committed to the Renshaw cell fate. This fate, they report, is determined by a temporal transcriptional program that involves selective activation of the Oc1 and Oc2 transcription factors during the first wave of V1 interneuron neurogenesis, broader expression of the Foxd3 transcription factor in postmitotic V1 interneurons, and later expression of the MafB transcription factor. Importantly, Renshaw cell specification and differentiation proceeds normally in the absence of neuronal activity. Overall, these results suggest that a combination of spatial and temporal determinants might be the major mechanism for generating neuronal diversity in the spinal cord.
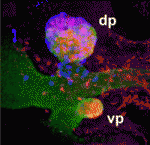
Topsy-turvy cilia precede neural progenitor delamination
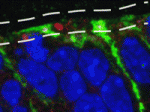
During the development of the mammalian cerebral cortex, basal progenitor cells delaminate from the apical adherens junction belt of the neuroepithelium. But what are the cell biological processes that precede the delamination of these neural progenitors? On p. 95, Wieland Huttner and co-workers identify a new pre-delamination state of neuroepithelial cells in the mouse embryonic neocortex. Using electron microscopy, the researchers show that, in a subpopulation of neuroepithelial cells, the re-establishment of the primary cilium after mitosis occurs at the basolateral rather than at the apical plasma membrane. Neuroepithelial cells carrying basolateral cilia selectively express the basal progenitor marker Tbr2, they report, and delaminate from the apical adherens junction belt to become basal progenitors. Notably, overexpression of insulinoma-associated 1, a transcription factor that promotes the generation of basal progenitors, increases the proportion of neuroepithelial cells with basolateral cilia. Together, these results suggest that the re-establishment of a basolateral primary cilium is a cell biological feature that precedes neural progenitor delamination.
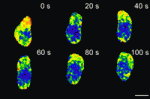
Digging into Polycomb repression
Polycomb group proteins control diverse developmental processes by repressing the transcription of developmental regulator genes through chromatin modification. The Drosophila Polycomb repressive complex 1 (PRC1), which contains the four core subunits Sce (Sex combs extra), Psc (Posterior sex combs), Ph (Polyhomeotic) and Pc (Polycomb), exhibits two chromatin-modifying activities – H2A monoubiquitylation and chromatin compaction. Here (p. 117), Jürg Müller and co-workers provide new insights into PRC1-mediated transcriptional repression. The researchers show that Sce is the major H2A ubiquitylase in Drosophila and identify the genes that are bound by PRC1. Then, by analysing mutants that lack individual PRC1 subunits, they identify two classes of PRC1 target genes. Repression of class I genes requires all four PRC1 core subunits whereas repression of class II genes requires only Psc and Ph. The researchers suggest, therefore, that H2A monoubiquitylation is crucial for the repression of a subset of PRC1 target genes but that chromatin compaction mediated by Psc and Ph may be the major mechanism by which PRC1 represses other target genes.
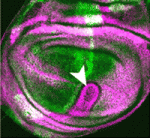
Gβγ signals polarise primordial germ cells
Directed cell migration occurs many times during embryogenesis. For example, in many species, primordial germ cells (PGCs) migrate after specification to the site of the future gonad. This migration involves PGC polarisation and PGC responsiveness to external guidance cues. In zebrafish, the chemokine Cxcl12a regulates directed migration, whereas the Rho GTPase Rac1 regulates polarisation. But what controls Rac activity? Fang Lin and co-workers now report that signalling mediated by the G protein subunits Gβ and Gγ (Gβγ) regulates Rac activity in zebrafish PGCs (see p. 57). The researchers show that PGCs defective for Gβγ signalling, like those with reduced Rac activity, fail to polarise and fail to migrate actively in response to directional cues. They also show that PGCs require Gβγ signalling for polarised activation of Rac and for maintenance of their overall Rac levels. These and other results suggest that, during PGC migration in zebrafish, Gβγ signalling regulates Rac activity to control the cell polarity that is needed for PGC responsiveness to chemokine signalling.
Plus…
Cell fate decisions and axis determination in the early mouse embryo
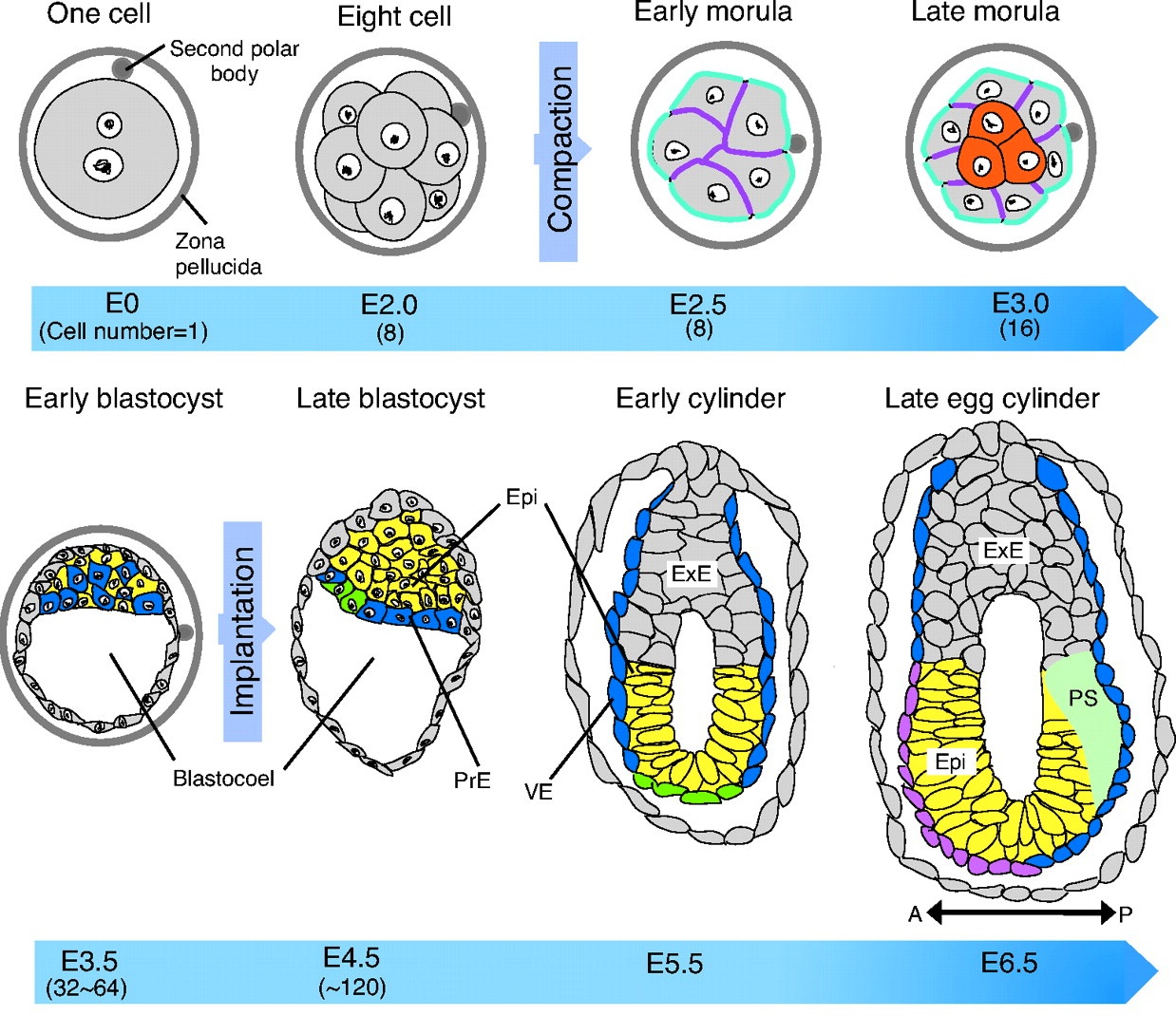
The mouse embryo generates multiple cell lineages, as well as its future body axes in the early phase of its development. Here, Takaoka and Hamada address the timing of the first cell fate decisions and of the establishment of embryonic polarity, and ask how far back one can trace their origins.
See the Review article on p. 3
Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells
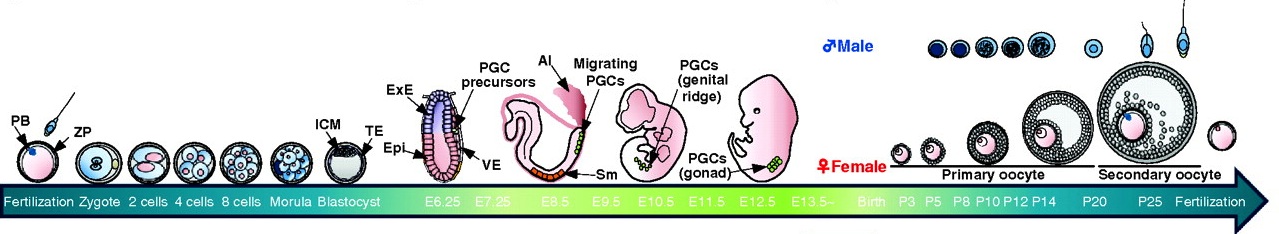
During mouse development, the epigenetic states of pre-implantation embryos and primordial germ cells undergo extensive reprogramming. Recent studies of DNA demethylation dynamics, reviewed by Saitou, Kagiwada and Kurimoto, provide new insights into the regulation of these epigenetic states. See the Review article on p. 15


 (No Ratings Yet)
(No Ratings Yet)