June in preprints
Posted by the Node, on 4 July 2018
Welcome to our monthly trawl for developmental biology (and other related) preprints!
This month we found a tranche of preprints getting deep into the mechanics of fly development, a clutch on organoids (retinal, cerebral, cortical!), an investigation into the role of gender in scientific collaboration, and a veritable zoo in our evo-devo section – from ladybirds to placozoans via pufferfish, hydra and choanoflagellates.
The preprints were hosted on bioRxiv, PeerJ, and arXiv. Let us know if we missed anything, and use these links to get to the section you want:
Developmental biology
| Stem cells, regeneration & disease modelling
Evo-devo & evo
Cell biology
Modelling
Tools & resources
Research practice & education
Why not…
Developmental biology
| Patterning & signalling
Canonical Wnt signaling regulates patterning, differentiation and nucleogenesis in mouse hypothalamus and prethalamus.
Elizabeth Newman, Dan Wu, Makoto Mark Taketo, Jiangyang Zhang, Seth Blackshaw
Dll1 and Jag1 are Differentially Required to Specify Proximal and Distal Pancreatic Duct Compartments
Philip A Seymour, Caitlin A Collin, Mette C Jorgensen, Itaru Imayoshi, Ryoichiro Kageyama, Palle Serup
Notch Controls Multiple Pancreatic Cell Fate Regulators Through Direct Hes1-mediated Repression
Kristian H de Lichtenberg, Philip A Seymour, Mette C Jorgensen, Yung-Hae Kim, Anne Grapin-Botton, Mark Magnuson, Nikolina Nakic, Jorge Ferrer, Palle Serup
Gain-of-function Shh mutants activate Smo in cis independent of Ptch1/2 function
Catalina Casillas, Henk Roelink
Sonic Hedgehog is a member of the Hh/DD-peptidase family that spans the eukaryotic and bacterial domains of life.
Henk Roelink
The chemokine receptor CXCR2 contributes to murine adipocyte development
Douglas P Dyer, Joan Boix Nebot, Christopher Kelly, Laura Medina-Ruiz, Fabian Scheutte, Gerard J Graham
Thyroid hormone signaling specifies cone subtypes in human retinal organoids
Kiara C Eldred, Sarah E Hadyniak, Katarzyna A Hussey, Boris Brennerman, Pingwu Zhang, Xitiz Chamling, Valentin M Sluch, Derek S Welsbie, Samer Hattar, James Taylor, Karl Wahlin, Donald J Zack, Robert J Johnston Jr.
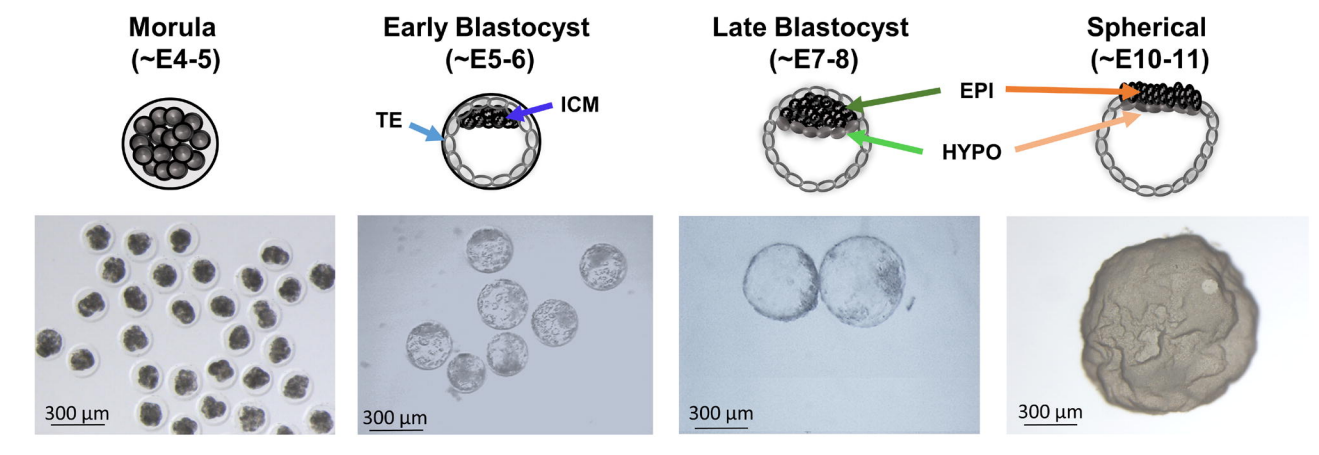
Lineage segregation, pluripotency and X-chromosome inactivation in the pig pre-gastrulation embryo
Priscila Ramos-Ibeas, Fei Sang, Qifan Zhu, Walfred W.C. Tang, Sarah Withey, Doris Klisch, Matthew Loose, Azim Surani, Ramiro Alberio
Chronic estrus disrupts uterine gland development and homeostasis
C. Allison Stewart, M. David Stewart, Ying Wang, Rui Liang, Yu Liu, Richard Behringer
Amot regulates neuronal dendritic tree through Yap1
Katarzyna O. Rojek, Joanna Krzemien, Hubert Dolezyczek, Pawel M. Boguszewski, Leszek Kaczmarek, Witold Konopka, Marcin Rylski, Jacek Jaworski, Lars Holmgren, Tomasz J. Proszynski
Single-cell ID-seq identifies BMP signaling as a driver of a late stage epidermal differentiation program.
Roderick E. van Eijl, Jessie A.G. van Buggenum, Sabine E.J. Tanis, Joost Hendriks, Klaas W. Mulder
Feedback Regulation between Initiation and Maturation Networks Orchestrates the Chromatin Dynamics of Epidermal Lineage Commitment
Lingjie Li, Yong Wang, Jessica L. Torkelson, Gautam Shankar, Jillian M. Pattison, Hanson H. Zhen, Zhana Duren, Fengqin Fang, Sandra P. Melo, Samantha N. Piekos, Jiang Li, Eric J. Liaw, Lang Chen, Rui Li, Marius Wernig, Wing H. Wong, Howard Y. Chang, Anthony E. Oro
Morphogen-Lineage Selector Interactions During Surface Epithelial Commitment
Sandra P Melo, Jillian M Pattison, Samantha N Piekos, Jessica L Torkelson, Elizaveta Bashkirova, Maxwell R Mumbach, Charlotte Rajasingh, Hanson Hui Zhen, Lingjie Li, Eric Liaw, Daniel Alber, Adam J Rubin, Gautam Shankar, Howard Y Chang, Paul A Khavari, Anthony E Oro
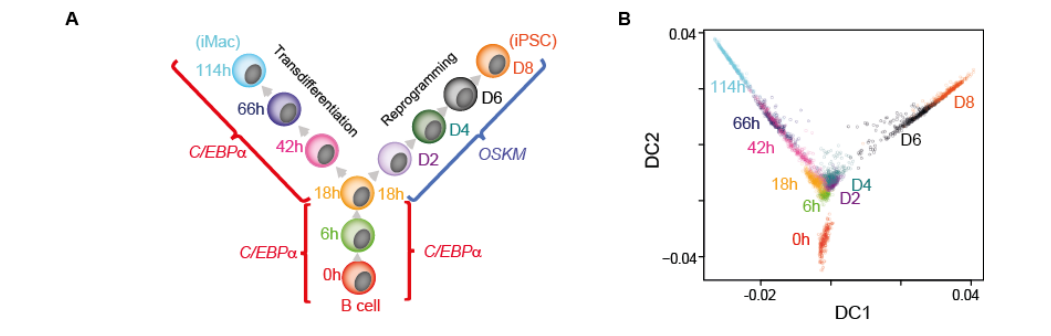
Single cell expression analysis uncouples transdifferentiation and reprogramming
Mirko Francesconi, Bruno Di Stefano, Clara Berenguer, Marisa de Andres, Maria Mendez Lago, Amy Guillaumet-Adkins, Gustavo Rodriguez-Esteban, Marta Gut, Ivo Gut, Holger Heyn, Ben Lehner, Thomas Graf
Rainbow-seq: combining cell lineage tracking with single-cell RNA sequencing in preimplantation embryos
Fernando Biase, Qiuyang Wu, Riccardo Calandrelli, Marcelo Rivas-Astroza, Shuigeng Zhou, Sheng Zhong
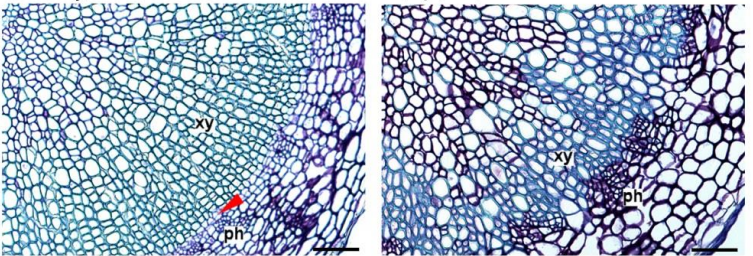
Paralogues of the PXY and ER receptor kinases enforce radial patterning in plant vascular tissue.
Ning Wang, Kristine S Bagdassarian, Rebecca Doherty, Xiao Wang, Johannes Kroon, Wei Wang, Ian Jermyn, Simon Turner, Peter Etchells
β-cyclocitral is a natural root growth regulator
Alexandra J Dickinson, Kevin Lehner, Medhavinee Mijar, Philip N Benfey
TTL proteins scaffold brassinosteroid signaling components at the plasma membrane to optimize signal transduction in plant cells
Vitor Amorim-Silva, Alvaro Garcia-Moreno, Araceli G Castillo, Naoufal Lakhssassi, Jessica Perez-Sancho, Yansha Li, Alicia Esteban del Valle, David Pose, Josefa Perez-Rodriguez, Jinxing Lin, Victoriano Valpuesta, Omar Borsani, Cyril Zipfel, Alberto P Macho, Miguel A Botella
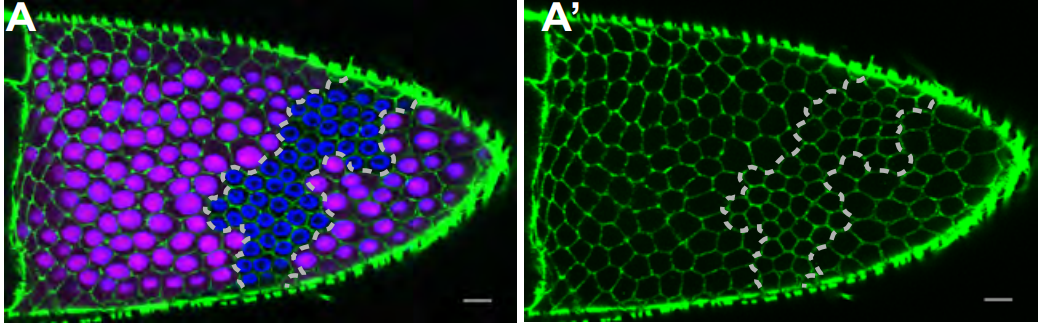
Drosophila IMP regulates Kuzbanian to control the timing of Notch signalling in the follicle cells
Weronicka Fic, Celia Faria, Daniel St Johnston
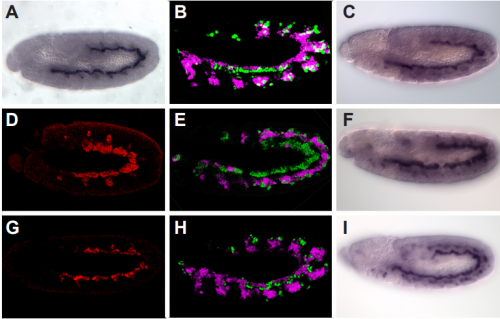
Distinct roles and requirements for Ras pathway signaling in visceral versus somatic muscle founder specification
Yiyun Zhou, Sarah E. Popadowski, Emily Deustchman, Marc S. Halfon
A repressor-decay timer for robust temporal patterning in embryonic Drosophila neuroblast lineages
Inna Averbukh, Sen-Lin Lai, Chris Q Doe, Naama Barkai
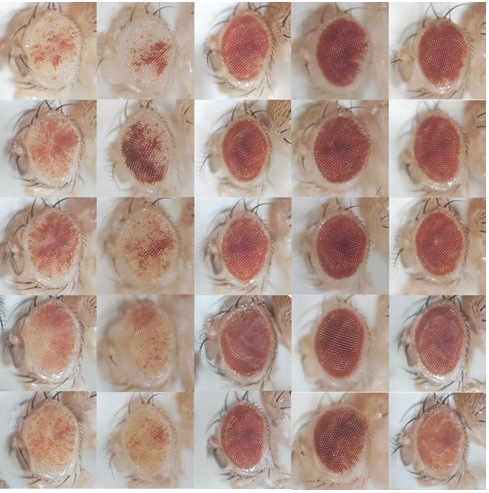
Altered hsrω lncRNA levels in activated Ras background further enhance Ras activity in Drosophila eye and induces more R7 photoreceptors
Mukulika Ray, Subhash C. Lakhotia
Signaling dynamics control cell fate in the early Drosophila embryo
Heath E Johnson, Stanislav Y Shvartsman, Jared E Toettcher
Kinetic sculpting of the seven stripes of the Drosophila even-skipped gene
Augusto Berrocal, Nicholas C Lammers, Hernan G Garcia, Michael B Eisen
A fat-tissue sensor couples growth to oxygen availability by remotely controlling insulin secretion
Michael J. Texada, Anne F. Joergensen, Daniel K. Smith, Dylan F.M. Marple, E. Thomas Danielsen, Sine K. Petersen, Jakob L. Hansen, Kenneth A. Halberg, Kim F. Rewitz
TORC1 modulation in adipose tissue is required for organismal adaptation to hypoxia in Drosophila.
Byoungchun Lee, Elizabeth C Barretto, Savraj S Grewal
Phosphatidylinositol 5 phosphate 4-kinase regulates insulin sensitivity and PIP3 turnover in Drosophila.
Sanjeev Sharma, Swarna Mathre, Ramya Visvanathan, Dhananjay Shinde, Padinjat Raghu
Glucose increases the lifespan of post-reproductive C. elegans independently of FOXO
Lei Wang, Caroline Beaudoin-Chabot, Guillaume Thibault
Perception of environmental polypeptides in C. elegans activates insulin/IGF signaling and alters lipid metabolism
Rebecca E. W. Kaplan, Amy K. Webster, Rojin Chitrakar, Joseph A. Dent, L. Ryan Baugh
A functional study of all 40 C. elegans insulin-like peptides
Shanqing Zheng, Hilton Chiu, Jeffrey Boudreau, Tony Papanicolaou, William Bendena, Ian Chin-Sang
Intergenerational effects of dietary restriction on insulin/IGF signaling and reproductive development
James M. Jordan, Jonathan D. Hibshman, Rebecca E. W. Kaplan, Amy K. Webster, Abigail Leinroth, Ryan Guzman, Colin S. Maxwell, Elizabeth Anne Bowman, E. Jane Albert Hubbard, L. Ryan Baugh
Variability in the timing of a β-catenin pulse biases a stochastic cell fate decision in C. elegans
Jason R Kroll, Jasonas Tsiaxiras, Jeroen S van Zon
| Morphogenesis & mechanics
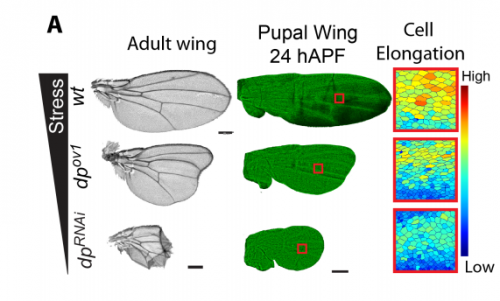
Mechanosensitive binding of p120-Catenin at cell junctions regulates E-Cadherin turnover and epithelial viscoelasticity
K. Venkatesan Iyer, Romina Piscitello-Gómez, Frank Jülicher, Suzanne Eaton
Analysis of the role of Nidogen/entactin in basement membrane assembly and morphogenesis in Drosophila
Maria Dolores Martin-Bermudo, Jianli Dai, Beatriz Estrada, Sofie Jacobs, Besaiz Sánchez-Sánchez, Jia Tang, Mengqi Ma, Patricia Magadan, José C. Pastor-Pareja
Characterization of Drosophila Nidogen/entactin reveals roles in basement membrane stability, barrier function and nervous system plasticity
Georg Wolfstetter, Ina Dahlitz, Kathrin Pfeifer, Joscha Arne Alt, Uwe Toepfer, Daniel Christoph Pfeifer, Reinhard Lakes-Harlan, Stefan Baumgartner, Ruth Palmer, Anne Holz
Dynamics of PAR proteins explain the oscillation and ratcheting mechanisms in dorsal closure
Clinton H. Durney, Tony J. C. Harris, James J. Feng
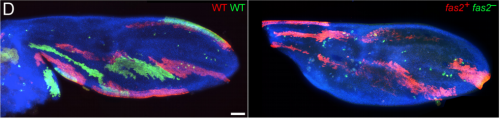
A Fasciclin 2 functional switch controls organ size in Drosophila
Emma Velasquez, Jose Gomez-Sanchez, Emmanuelle Donier, Carmen Grijota-Martinez, Hugo Cabedo, Luis A Garcia-Alonso
Targeted substrate degradation by Kelch controls the actin cytoskeleton during ring canal expansion
Andrew M. Hudson, Katelynn M. Mannix, Julianne A. Gerdes, Molly C. Kottemann, Lynn Cooley
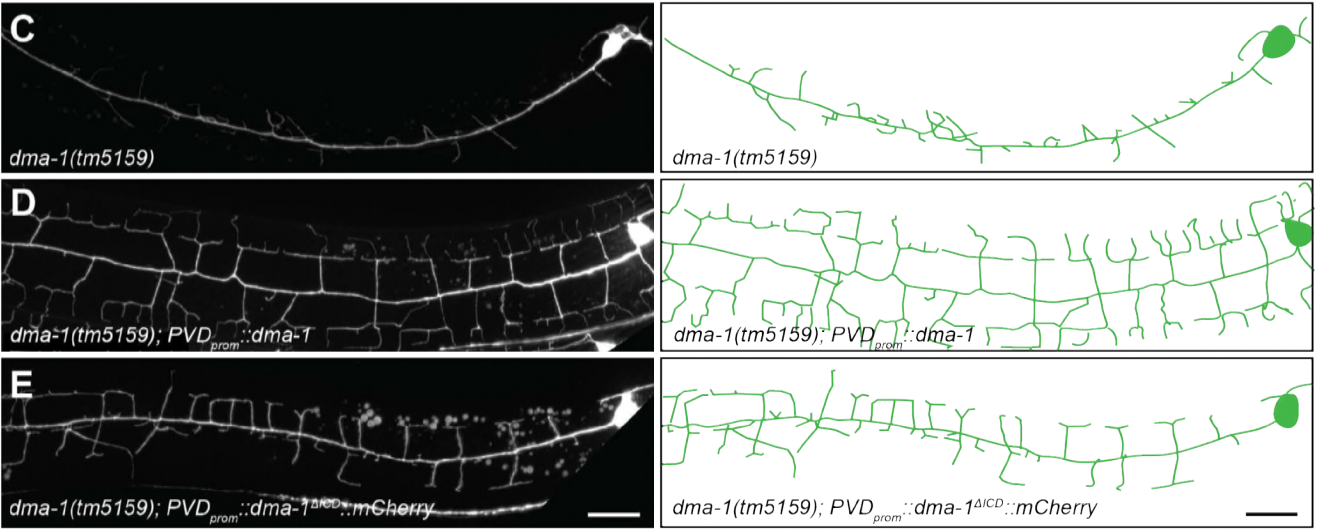
TIAM-1/GEF can shape somatosensory dendrites independently of its GEF activity by regulating F-actin localization
Leo T.H. Tang, Carlos A Diaz-Balzac, Maisha Rahman, Nelson J Ramirez-Suarez, Yehuda Salzberg, Maria I Lazaro-Pena, Hannes E Bülow
Distinct CED-10/Rac1 Domains Confer Context-Specific Functions in Neuronal Development
Roger Pocock, Steffen Noergaard, Shuer Deng, Wei Cao
Novel exc Genes Involved in Formation of the Tubular Excretory Canals of C. elegans
Hikmat Al-Hashimi, Travis Chiarelli, Erik A. Lundquist, Matthew Buechner
Size control of the inner ear via hydraulic feedback
Kishore R. Mosaliganti, Ian A. Swinburne, Chon U Chan, Nikolaus D. Obholzer, Amelia A. Green, Shreyas Tanksale, L Mahadevan, Sean G. Megason
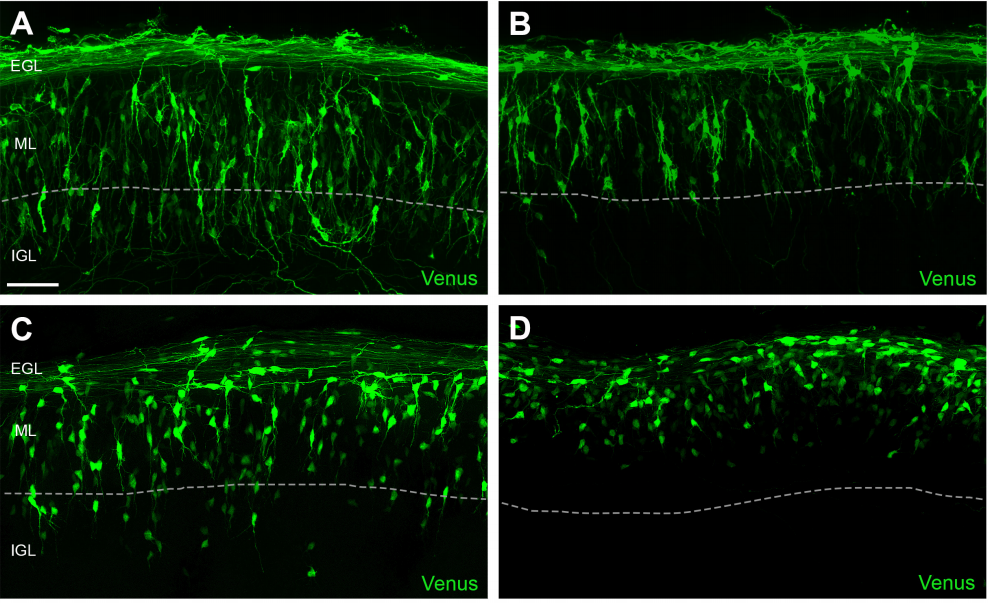
N-Cadherin Provides a Cis and Trans Ligand for Astrotactin that Functions in Glial-Guided Neuronal Migration
Zachi Horn, Hourinaz Behesti, Mary E. Hatten
Actomyosin dynamics, Bmp and Notch signaling pathways drive apical extrusion of proepicardial cells
Laura Andres-Delgado, Alexander Ernst, Maria Galardi-Castilla, David Bazaga, Marina Peralta Lopez, Juliane Munch, Juan Manuel Gonzalez-Rosa, Federico Tessadori, Jeroen Bakkers, Jose Luis De La Pompa Minguez, Julien Vermot, Nadia Mercader
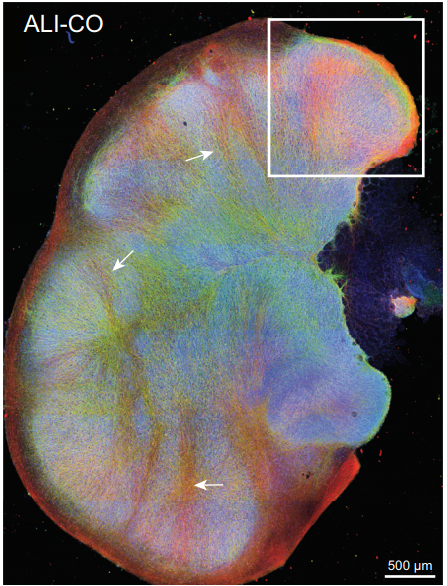
Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output
Stefano L Giandomenico, Susanna B Mierau, George M Gibbons, Lea MD Wenger, Laura Masullo, Timothy Sit, Magdalena Sutcliffe, Jerome Boulanger, Marco Tripodi, Emmanuel Derivery, Ole Paulsen, Andras Lakatos, Madeline Lancaster
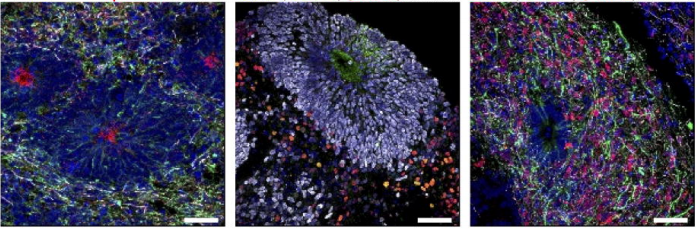
Nested oscillatory dynamics in cortical organoids model early human brain network development
Cleber A. Trujillo, Richard Gao, Priscilla D. Negraes, Isaac A. Chaim, Alain Domissy, Matthieu Vandenberghe, Anna Devor, Gene W. Yeo, Bradley Voytek, Alysson R. Muotri
Cerebellar nuclei neurons dictate growth of the cortex through developmental scaling of presynaptic Purkinje cells
Ryan T Willett, Alexandre Wojcinski, Nermin Sumru Bayin, Zhimin Lao, Daniel Stephen, Katherine L Dauber-Decker, Zhuhao Wu, Marc Tessier-Lavigne, Hiromitsu Saito, Noboru Suzuki, Alexandra L Joyner
Three-dimensional organization of transzonal projections and other cytoplasmic extensions in mouse ovarian follicles
Valentina Baena, Mark Terasaki
Active fluctuations modulate gene expression in mouse oocytes
Maria Almonacid, Stephany El-Hayek, Alice Othmani, Isabelle Queguiner, Fanny Coulpier, Sophie Lemoine, Leïla Bastianelli, Christophe Klein, Tristan Piolot, Philippe Mailly, Raphaël Voituriez, Auguste Genovesio, Marie-Hélène Verlhac
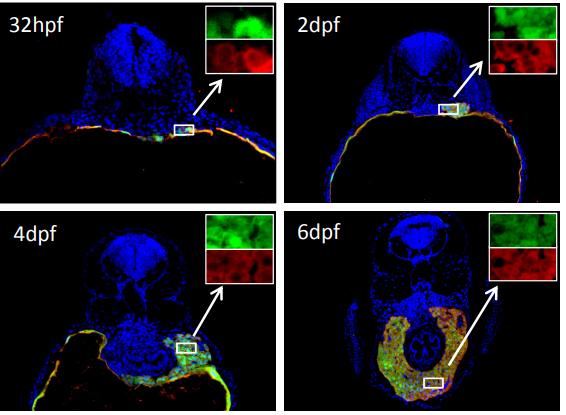
Zebrafish hhex null mutant develops an intrahepatic intestinal tube due to de-repression of cdx1b and pdx1
Ce Gao, Weidong Huang, Yuqi Gao, Li Jan Lo, Lingfei Luo, Honghui Huang, Jun Chen, Jinrong Peng
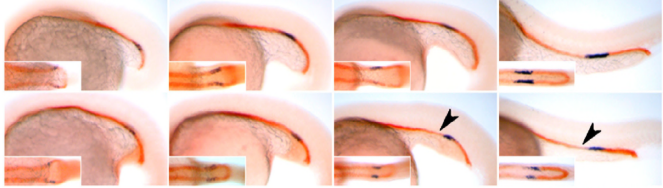
A novel mechanism of gland formation in zebrafish involving transdifferentiation of renal epithelial cells and live cell extrusion
Richard W Naylor, Alan J Davidson
Linear stability analysis of morphodynamics during tissue regeneration in plants
Anne-Mieke Reijne, Gunnar Pruessner, Giovanni Sena
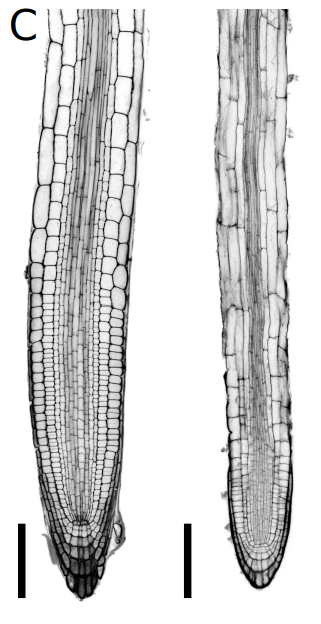
Light dynamically regulates growth rate and cellular organisation of the Arabidopsis root meristem.
Thomas Blein, Jasmin Duerr, Taras Pasternak, Thomas Haser, Thorsten Falk, Kun Liu, Franck Anicet Ditengou, Olaf Ronneberger, Klaus Palme
Cellular heterogeneity in pressure and growth emerges from tissue topology and geometry
Yuchen Long, Ibrahim Cheddadi, Vincent Mirabet, Mathilde Dumond, Christophe Godin, Arezki Boudaoud
| Genes & genomes
Pou5f3, SoxB1 and Nanog remodel chromatin on High Nucleosome Affinity Regions at Zygotic Genome Activation
Marina Veil, Lev Yampolsky, Bjoern Gruening, Daria Onichtchouk
Nucleoporin 133 deficiency leads to glomerular damage in zebrafish
Chiara Cianciolo Cosentino, Alessandro Berto, Michelle Hari, Johannes Loffing, Stephan C. F. Neuhauss, Valerie Doye
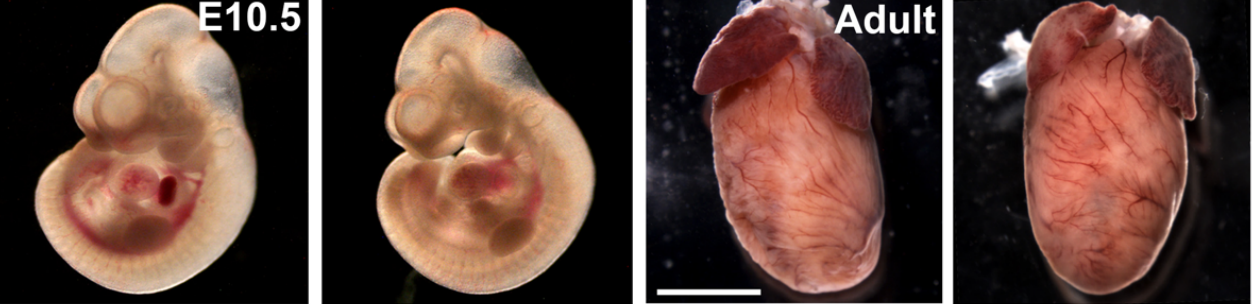
Myc is dispensable for cardiac development in the mouse but can rescue Mycn-deficient hearts through functional replacement and cell competition
Noelia Muñoz-Martín, Rocío Sierra, Thomas Schimmang, Cristina Villa del Campo, Miguel Torres
Timed collinear activation of Hox genes during gastrulation controls the avian forelimb position
Chloe Moreau, Paolo Caldarelli, Didier Rocancourt, Julian Roussel, Nicolas Denans, Olivier Pourquie, Jerome Gros
Tbx1 represses Mef2c gene expression by inducing histone 3 deacetylation of the anterior heart field enhancer
Luna Simona Pane, Filomena Gabriella Fulcoli, Andrea Cirino, Alessandra Altomonte, Rosa Ferrentino, Marchesa Bilio, Antonio Baldini
DNMT1 in Six2 progenitor cells is essential for transposable element silencing and kidney development
Szu-Yuan Li, Jihwan Park, Kiwung Chung, Rojesh Shrestha, Matthew B Palmer, Katalin Susztak
Using human sequencing to guide craniofacial research.
Ryan Liegel, Erin Finnerty, Lauren Ward, Andrew DiStasio, Robert Hufnagel, Howard Saal, Cynthia Prows, Rolf Stottmann, Rolf Stottmann
Spatiotemporal Gene Coexpression and Regulation in Mouse Cardiomyocytes of Early Cardiac Morphogenesis
Yang Liu, Pengfei Lu, Yidong Wang, Bernice E. Morrow, Bin Zhou, Deyou Zheng
Genome-wide analysis of H3K4me3 and H3K27me3 modifications throughout the mouse urogenital ridge at E11.5.
Yisheng Yang, Megan J Wilson
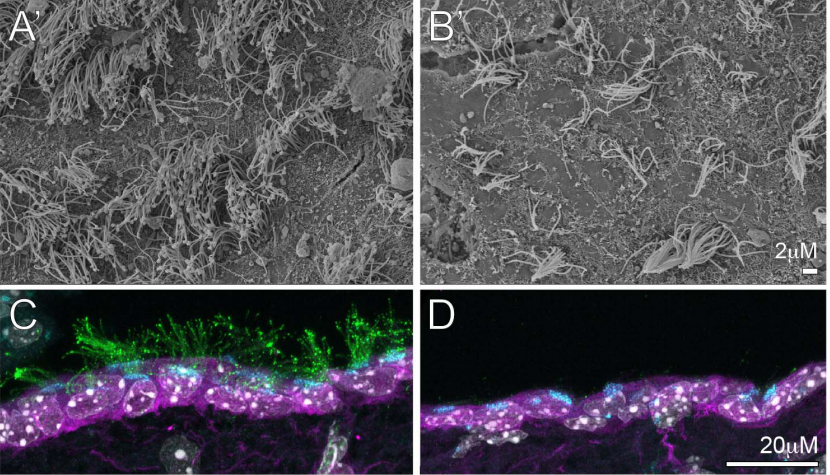
Mutations in Kinesin Family Member 6 Reveal Specific Role in Ependymal Cell Function and Human Neuro-Cranial Development
Mia J Konjikusic, Patra Yeetong, Rungnapa Ittiwut, Kanya Suphapeetiporn, John B Wallingford, Christina A Gurnett, Vorasuk Shotelersuk, Ryan Gray
Cardiac differentiation at single cell resolution reveals a requirement of hypertrophic signaling for HOPX transcription
Clayton Friedman, Quan Nguyen, Samuel Lukowski, Abbigail Helfer, Han Chiu, Holly Voges, Shengbao Suo, Jing-Dong Han, Pierre Osteil, Guangdun Peng, Naihe Jing, Greg Ballie, Anne Senabouth, Angelika Christ, Timothy Bruxner, Charles Murry, Emily Wong, Jun Ding, Yuliang Wang, James Hudson, Ziv Bar-Joseph, Patrick Tam, Joseph Powell, Nathan Palpant
Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis
Christina Ernst, Nils Eling, Celia P. Martinez-Jimenez, John C. Marioni, Duncan T. Odom
Altering the temporal regulation of one transcription factor drives sensory trade-offs
Ariane Ramaekers, Simon Weinberger, Annelies Claeys, Martin Kapun, Jiekun Yan, Reinhard Wolf, Thomas Flatt, Erich Buchner, Bassem A Hassan
The BTB-ZF transcription factor Tramtrack 69 shapes neural cell lineages by coordinating cell proliferation and cell fate
Francoise Simon, Anne Ramat, Sophie Louvet-Vallee, Angelique Burg, Agnes Audibert, Michel Gho
Drosophila small ovary encodes a zinc-finger repressor required for ovarian differentiation
Leif Benner, Elias A Castro, Cale Whitworth, Koen JT Venken, Haiwang Yang, Brian Oliver, Dorothy Lerit, Kevin R Cook
The mirtron miR-1010 functions in concert with its host gene SKIP to maintain synaptic homeostasis
Christopher Amourda, Timothy E Saunders
Contrasting effects of aging on the expression of transposons, the piRNA machinery and mitochondrial transcripts in the Drosophila ovary.
Alexandra A. Erwin, Justin P. Blumenstiel
Gene-regulatory independent functions for insect DNA methylation
Adam J. Bewick, Zachary Sanchez, Elizabeth C. Mckinney, Allen J. Moore, Patricia J. Moore, Robert J. Schmitz
Chromatin accessibility is dynamically regulated across C. elegans development and ageing
Jurgen Janes, Yan Dong, Michael Schoof, Jacques Serizay, Alex Appert, Chiara Cerrato, Carson Woodbury, Ron Chen, Carolina Gemma, Ni Huang, Djem Kissiov, Przemyslaw Stempor, Annette Steward, Eva Zeiser, Sascha Sauer, Julie Ahringer
Regulated nuclear accumulation of a histone methyltransferase times the onset of heterochromatin formation in C. elegans embryos
Beste Mutlu, Huei-Mei Chen, James J. Moresco, Barbara D. Orelo, Bing Yang, John M. Gaspar, Sabine Keppler-Ross, John R. Yates III, David H. Hall, Eleanor M. Maine, Susan E. Mango
ZAG-1/ZEB prevents the expression of repressors and protects neuronal cell fate in Caenorhabditis elegans
Chaogu Zheng, Felix Qiaochu Jin, Brian Loeber Trippe, Ji Wu, Martin Chalfie
H3K9me3 is Required for Transgenerational Inheritance of Small RNAs that Target a Unique Subset of Newly Evolved Genes
Itamar Lev, Hila Gingold, Oded Rechavi
Dauer diapause has transgenerational effects on starvation survival and gene expression plasticity
Amy K Webster, James M Jordan, Jonathan D Hibshman, Rojin Chitrakar, L Ryan Baugh
Spatial transcriptomics of C. elegans males and hermaphrodites identifies novel fertility genes
Annabel Ebbing, Abel Vertesy, Marco Betist, Bastiaan Spanjaard, Jan Philipp Junker, Alexander van Oudenaarden, Eugene Berezikov, Hendrik Korswagen
A variably imprinted epiallele impacts seed development
Daniela Pignatta, Katherine Novitzky, P.R. V. Satyaki, Mary Gehring
Developmental genetics of corolla tube formation: role of the tasiRNA-ARF pathway
Baoqing Ding, Rui Xia, Qiaoshan Lin, Vandana Gurung, Janelle M. Sagawa, Lauren E. Stanley, Matthew Strobel, Pamela K. Diggle, Blake C. Meyers, Yao-Wu Yuan
Distinct types of short open reading frames are translated in plant cells
Igor Fesenko, Ilya Kirov, Andrey Kniazev, Regina Khazigaleeva, Vassili Lazarev, Daria Kharlampieva, Ekaterina Grafskaia, Viktor Zgoda, Ivan Butenko, Georgy Arapidi, Anna Mamaeva, Vadim Ivanov, Vadim Govorun
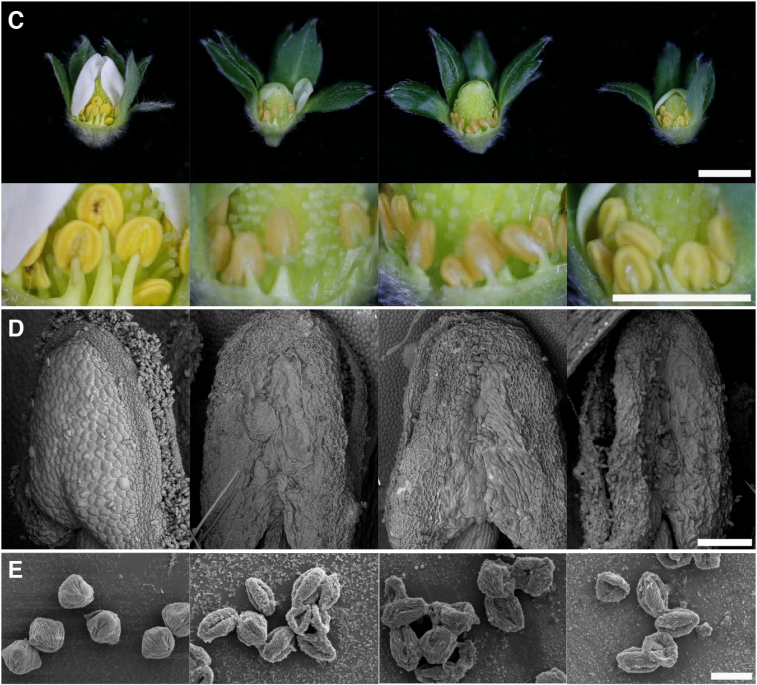
Functional Analysis of TM6 MADS box gene in the Octoploid Strawberry by CRISPR/Cas9 directed mutagenesis
Carmen Martín-Pizarro, Juan Carlos Triviño, David Posé
Exploration of miRNA-mediated fertility regulation network of cytoplasmic male sterility during flower bud development in soybean
Xianlong Ding, Hao Zhang, Hui Ruan, Yanwei Li, Linfeng Chen, Tanliu Wang, Ling Jin, Xiaoqiang Li, Shouping Yang, Junyi Gai
A Genome-Wide Association Study Reveals a Novel Regulator of Ovule Number and Fertility in Arabidopsis thaliana
Jing Yuan, Sharon A Kessler
| Stem cells, regeneration & disease modelling
Conservation of epigenetic regulation by the MLL3/4 tumour suppressor in planarian pluripotent stem cells
Yuliana Mihaylova, Prasad Abnave, Damian Kao, Samantha Hughes, Alvina Lai, Farah Jaber-Hijazi, Nobuyoshi Kosaka, Aziz Aboobaker
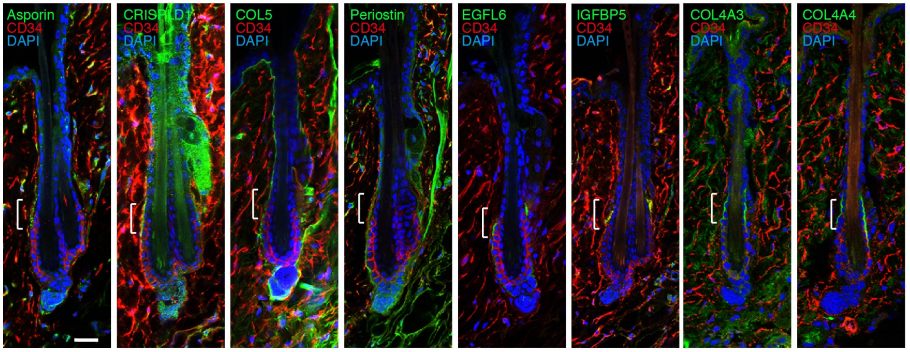
Hair Follicle Epidermal Stem Cells Define a Niche for Tactile Sensation
Chun-Chun Cheng, Ko Tsutsui, Toru Taguchi, Noriko Sanzen, Asako Nakagawa, Kisa Kakiguchi, Shigenobu Yonemura, Chiharu Tanegashima, Sean D Keeley, Hiroshi Kiyonari, Yasuhide Furuta, Yasuko Tomono, Fiona M. Watt, Hironobu Fujiwara
“Ancestralization” of human pluripotent stem cells by multiplexed precise genome editing
Stephan Riesenberg, Tomislav Maricic, Svante Pääbo
A common molecular logic determines embryonic stem cell self-renewal and reprogramming
Sara-Jane Dunn, Meng Amy Li, Elena Carbognin, Austin G Smith, Graziano Martello
Single-cell transcriptomics identifies CD44 as a new marker and regulator of haematopoietic stem cells development
Morgan Oatley, Ozge Vargel Bolukbasi, Valentine Svensson, Maya Shvartsman, Kerstin Ganter, Katharina Zirngibl, Polina V Pavlovich, Vladislava Milchevskaya, Vladimira Foteva, Kedar N. Natarajan, Bianka Baying, Vladimir Benes, Kiran Raosaheb Patil, Sarah A. Teichmann, Christophe Lancrin
LncRNA Spehd regulates hematopoietic stem cells and progenitors and is required for multilineage differentiation
M Joaquina Delás, Benjamin T Jackson, Tatjana Kovacevic, Silvia Vangelisti, Ester Munera Maravilla, Sophia A Wild, Eva Maria Stork, Nicolas Erard, Simon RV Knott, Gregory J Hannon
O-GlcNAc Homeostasis Controls Cell Fate Decisions During Hematopoiesis
Zhen Zhang, Matt P Parker, Stefan Graw, Lesya Novikova, Halyna Fedosyuk, Joseph Fontes, Devin Koestler, Kenneth Peterson, Chad Slawson
Histone Acetyltransferase Kat2a Stabilises Pluripotency with Control of Transcriptional Heterogeneity
Naomi Moris, Shlomit Edri, Denis Seyres, Rashmi Kulkarni, Ana Filipa Domingues, Tina Balayo, Mattia Frontini, Cristina Pina
Profiling of pluripotency factors in individual stem cells and early embryos
Sarah J Hainer, Ana Boskovic, Oliver J Rando, Thomas G Fazzio
Genome-scale oscillations in DNA methylation during exit from pluripotency
Steffen Rulands, Heather J Lee, Stephen J Clark, Christof Angermueller, Sebastien A Smallwood, Felix Krueger, Hisham Mohammed, Wendy Dean, Jennifer Nichols, Peter Rugg-Gunn, Gavin Kelsey, Oliver Stegle, Benjamin D Simons, Wolf Reik
Epigenetic-scale comparison of human iPSCs generated by retrovirus, Sendai virus or episomal vectors
Koichiro Nishino, Yoshikazu Arai, Ken Takasawa, Masashi Toyoda, Mayu Yamazaki-Inoue, Tohru Sugawara, Hidenori Akutsu, Ken Nishimura, Manami Ohtaka, Mahito Nakanishi, Akihiro Umezawa
Knockdown of butyrylcholinesterase but not inhibition by chlorpyrifos alters early differentiation mechanisms in human neural stem cells
Angela K. Teithof, Jason R. Richardson, Ronald P. Hart
In situ transcriptome characteristics are lost following culture adaptation of adult cardiac stem cells
Taeyong Kim, Oscar H Echeagaray, Bingyan J Wang, Alexandria Casillas, Kathleen M. Broughton, Bong-Hyun Kim, Mark A Sussman
Insights into the mutational burden of human induced pluripotent stem cells using an integrative omics approach
Matteo D’Antonio, Paola Benaglio, David A. Jakubosky, William W. Greenwald, Hiroko Matsui, Margaret K.R. Donovan, He Li, Erin N. Smith, Agnieszka D’Antonio-Chronowska, Kelly A. Frazer
miR17~92 is essential for the survival of hematopoietic stem and progenitor cells by restraining pro-apoptotic BIM
Kerstin Brinkmann, Craig Hyland, Carolyn A de Graaf, Andreas Strasser, Warren S Alexander, Marco J Herold
Interrupted reprogramming into induced pluripotent stem cells does not rejuvenate human mesenchymal stromal cells
Carolin Goebel, Roman Goetzke, Thomas Eggermann, Wolfgang Wagner
Muscle Stem Cell Niche Dysregulation in Volumetric Muscle Loss Injury
Shannon Anderson, Woojin M Han, Vunya Srinivasa, Mahir Mohiuddin, Marissa A Ruehle, Austin Moon, Eunjung Shin, Cheryl L San Emeterio, Molly E Ogle, Edward A Botchwey, Nick J Willett, Young C Jang
Effective repair of joint cartilage using human pluripotent stem cell-derived tissue
Oliver F.W. Gardner, Subhash Juneja, Heather Whetstone, Yulia Nartiss, Jakob T Sieker, Christian Veillette, Gordon M Keller, April M Craft
Functional Role of Annexins in Zebrafish Caudal Fin Regeneration: A Gene Knockdown Approach in Regenerating Tissue.
Mir Quoseena, Sowmya Vuppaladadium, Shahid Hussain, Swarna Bharathi, Mohammed M Idris
Regulatory T-cells are required for neonatal heart regeneration
Jiatao Li, Kevin Y Yang, Rachel Chun Yee Tam, Vicken W Chan, Bao Sheng, Shohei Hori, Bin Zhou, Yuk Ming Dennis Lo, Kathy O Lui
RAB-5 regulates regenerative axonal fusion by controlling EFF-1 endocytosis
Casey Linton, Brent Neumann, Rosina Giordano-Santini, Massimo A Hilliard
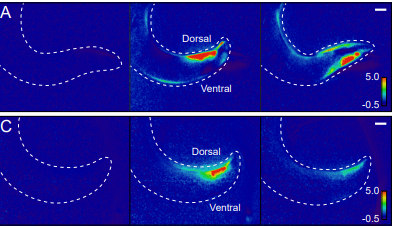
Aberrant Information Transfer Interferes with Functional Axon Regeneration
Chen Ding, Marc Hammarlund
Combined inhibition of apoptosis and necrosis promotes transient neuroprotection of retinal ganglion cells and partial axon regeneration after optic nerve damage
Maki Kayama, Kumiko Omura, Yusuke Murakami, Edith Reshef, Aristomenis Thanos, Yuki Morizane, Andrea Gianni, Toru Nakazawa, Joan Miller, Larry Benowitz, Demetrios Vavvas
Cerebrovascular damages induce lymphatic invasion into brain parenchyma to guide vascular regeneration
Jingying Chen, Jianbo He, Qifen Yang, Yaoguang Zhang, Tianyu Zhao, Deqin Yang, Lingfei Luo
The Drosophila SWI/SNF chromatin-remodeling complexes play separate roles in regulating growth and cell fate during regeneration
Yuan Tian, Rachel K Smith-Bolton
Functional testing of a human PBX3 variant in zebrafish reveals a potential modifier role in congenital heart defects
Gist H. Farr III, Kimia Imani, Darren Pouv, Lisa Maves
Shared gene co-expression networks in autism from induced pluripotent stem cell (iPSC) neurons
Dwaipayan Adhya, Vivek Swarup, Paulina Nowosaid, Carole Shum, Kamila Jozwik, Grainne McAlonan, Maria Andreina Mendez, Jamie Horder, Declan Murphy, Daniel H. Geschwind, Jack Price, Jason Carroll, Deepak P. Srivastava, Simon Baron-Cohen
Complete Disruption of Autism-Susceptibility Genes by Gene-Editing Predominantly Reduces Functional Connectivity of Isogenic Human Neurons
Eric Deneault, Sean H White, Deivid C Rodrigues, Joel Ross, Muhammad Faheem, Kirill Zaslavsky, Zhuozhi Wang, Roumiana Alexandrova, Giovanna Pellecchia, Wei Wei, Alina Piekna, Gaganjot Kaur, Jennifer L Howe, Vickie Kwan, Bhooma Thiruvahindrapuram, Susan Walker, Peter Pasceri, Daniele Merico, Ryan KC Yuen, Karun K Singh, James Ellis, Stephen W Scherer
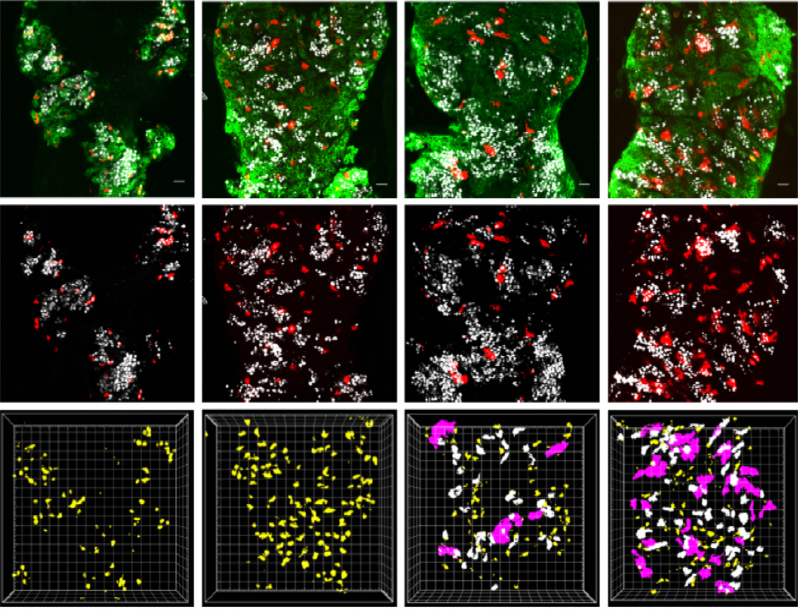
Cooption of antagonistic RNA-binding proteins establishes cell hierarchy in Drosophila neuro-developmental tumors
Sara Genovese, Raphael Clement, Cassandra Gaultier, Florence Besse, Karine Narbonne-Reveau, Fabrice Daian, Sophie Foppolo, Nuno Miguel Luis, Cédric Maurange
Vascular defects of DYRK1A knockouts are ameliorated by modulating calcium signaling in zebrafish
Jeong-Soo Lee, Hyun-Ju Cho, Jae-Geun Lee, Jong-Hwan Kim, Seon-Young Kim, Yang Hoon Huh, Hyo-Jeong Kim, Kyu-Sun Lee, Kweon Yu
Developmental-stage-specific proliferation and retinoblastoma genesis in RB-deficient human but not mouse cone precursors
Hardeep P Singh, Sijia Wang, Kevin Stachelek, Sunhye Lee, Mark W Reid, Matthew E Thornton, Cheryl Mae Craft, Brendan H Grubbs, David Cobrinik
The pyriproxyfen metabolite 4’OH- pyriproxyfen disrupts thyroid hormone signaling and enhances Musashi-1 levels in neuroprogenitors.
Petra Spirhanzlova, Sebastien Le Mevel, Karn Wejaphikul, Bilal Mughal, Jean-David Gothie, Anthony Sebillot, Lucille Butruille, Michelle Leemans, Theo Visser, Sylvie Remaud, Jean-Baptiste Fini, Barbara Demeneix
Defining Hepatic Modifiers Of ATTR Amyloidosis Through Therapeutic Gene Editing In Patient iPSCs
Richard M Giadone, Derek C Liberti, Taylor M Matte, Nicholas Skvir, Kai-Chun Chen, Jyh-Chang Jean, Andrew A Wilson, Darrell N Kotton, R Luke Wiseman, George J Murphy
Evo-devo & evo
The Ly6/uPAR protein Bouncer is necessary and sufficient for species-specific fertilization
Sarah Herberg, Krista R Gert, Alexander Schleiffer, Andrea Pauli
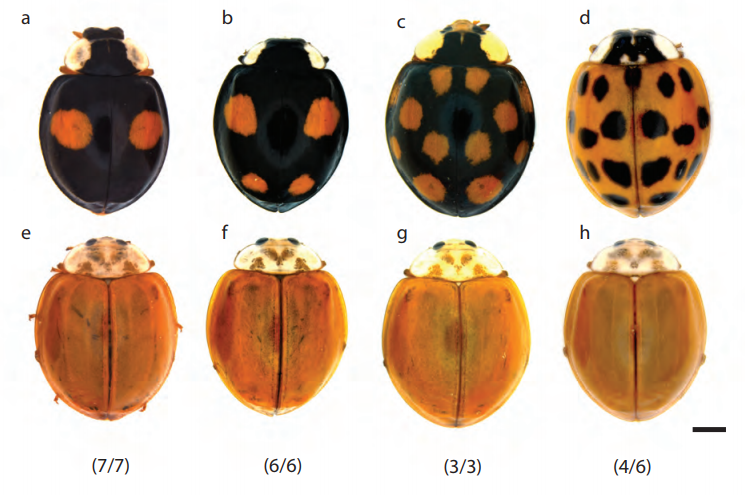
The genomic basis of colour pattern polymorphism in the harlequin ladybird
Mathieu Gautier, Junichi Yamaguchi, Julien Foucaud, Anne Loiseau, Aurelien Ausset, Benoit Facon, Bernhard Gschloessl, Jacques Lagnel, Etienne Loire, Hugues Parrinello, Danny Severac, Celine Lopez-Roques, Cecile Donnadieu, Maxime Manno, Helene Berges, Karim Gharbi, Lori Lawson-Handley, Lian-Sheng Zang, Heiko Vogel, Arnaud Estoup, Benjamin Prud’homme
Repeated inversions at the pannier intron drive diversification of intraspecific colour patterns of ladybird beetles
Toshiya Ando, Takeshi Matsuda, Kumiko Goto, Kimiko Hara, Akinori Ito, Junya Hirata, Joichiro Yatomi, Rei Kajitani, Miki Okuno, Katsushi Yamaguchi, Masaaki Kobayashi, Tomoyuki Takano, Yohei Minakuchi, Masahide Seki, Yutaka Suzuki, Kentaro Yano, Takehiko Itoh, Shuji Shigenobu, Atsushi Toyoda, Teruyuki Niimi
A revised understanding of Tribolium morphogenesis further reconciles short and long germ development
Matthew Alan Benton
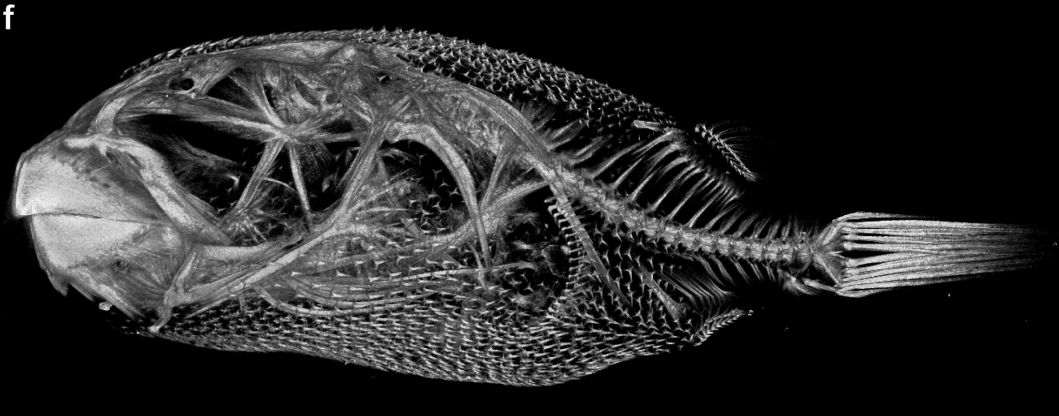
Evolution and developmental diversity of skin spines in pufferfish
Takanori Shono, Alexandre P Thiery, Daisuke Kurokawa, Ralf Britz, Gareth J Fraser
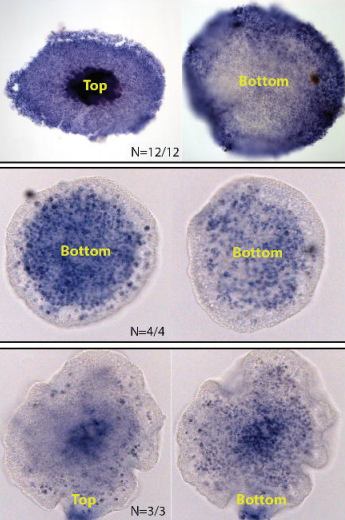
The radial expression of dorsal-ventral patterning genes in placozoans, Trichoplax adhaerens, argues for an oral-aboral axis
Timothy DuBuc, Yuriy Bobkov, Joseph F Ryan, Mark Martindale
CRISPR/Cas9-mediated gene knockin in the hydroid Hydractinia symbiolongicarpus
Steven M Sanders, Zhiwei Ma, Julia M Hughes, Brooke M Riscoe, Gregory A Gibson, Alan M Watson, Hakima Flici, Uri Frank, Christine E Schnitzler, Andreas D Baxevanis, Matthew L Nicotra
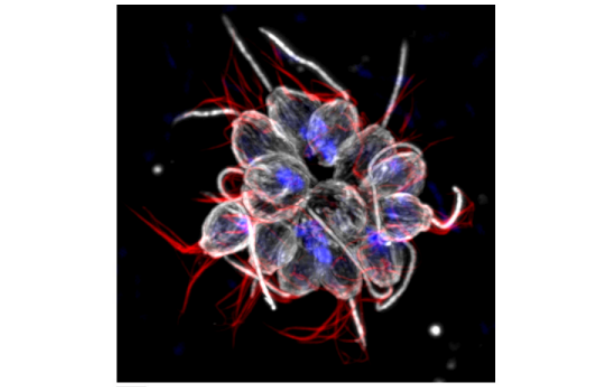
A robust method for transfection in choanoflagellates illuminates their cell biology and the ancestry of animal septins
David Booth, Heather Middleton, Nicole King
Transcriptomic atlas of mushroom development highlights an independent origin of complex multicellularity
Krisztina Krizsan, Eva Almasi, Zsolt Merenyi, Neha Sahu, Mate Viragh, Tamas Koszo, Stephen Mondo, Brigitta Kiss, Balazs Balint, Ursula Kues, Kerrie Barry, Judit Cseklye, Botond Hegedus, Bernard Henrissat, Jenifer Johnson, Anna Lipzen, Robin A. Ohm, Istvan Nagy, Jasmyn Pangilinan, Juying Yan, Yi Xiong, Igor V. Grigoriev, David S. Hibbett, Laszlo G. Nagy
Evolutionary trade-offs between unicellularity and multicellularity in budding yeast
Jennie Kuzdzal-Fick, Lin Chen, Gabor Balazsi
De novo origin of multicellularity in response to predation
Matthew D. Herron, Josh M. Borin, Jacob C. Boswell, Jillian Walker, I-Chen Kimberly Chen, Charles A. Knox, Margrethe Boyd, Frank Rosenzweig, William C. Ratcliff
Morphological, cellular and molecular characterization of posterior regeneration in the marine annelid Platynereis dumerilii
Anabelle Planques, Julien Malem, Julio Parapar, Michel Vervoort, eEve Gazave
Miniscule differences between the sex chromosomes in the giant genome of a salamander, Ambystoma mexicanum
Melissa Carol Keinath, Nataliya Timoshevskaya, Vladimir A Timoshevskiy, S. Randal Voss, Jeramiah J Smith
Evolution of salivary glue genes in Drosophila species
Jean-Luc Da Lage, Gregg W. C. Thomas, Magalie Bonneau, Virginie Courtier-Orgogozo
Diet-based assortative mating through sexual imprinting
Emily K. Delaney, Hopi E. Hoekstra
Chromosomal rearrangements as a source of new gene formation in Drosophila yakuba
Nicholas B. Stewart, Rebekah L. Rogers
Medaka population genome structure and demographic history described via genotyping-by-sequencing
Takafumi Katsumura, Shoji Oda, Hiroshi Mitani, Hiroki Oota
Architectural traits constrain the evolution of unisexual flowers and sexual segregation within inflorescences: an interspecific approach
Ruben Torices, Ana Afonso, Arne A. Anderberg, Jose M. Gomez, Marcos Mendez
Cell biology
Separation and Loss of Centrioles from Primordidal Germ Cells to Mature Oocytes in the Mouse
Gerald Schatten, Calvin R Simerly, Marion Manil-Ségalen, Carlos Castro, Carrie Hartnett, Dong Kong, Marie-Helene Verlhac, Jadranka Loncarek
Nuclear actin is required for transcription during Drosophila oogenesis
Maria Sokolova, Henna M Moore, Bina Prajapati, Joseph Dopie, Leena Merilainen, Mikko Honkanen, Rita Cerejeira Matos, Minna Poukkula, Ville Hietakangas, Maria Vartiainen
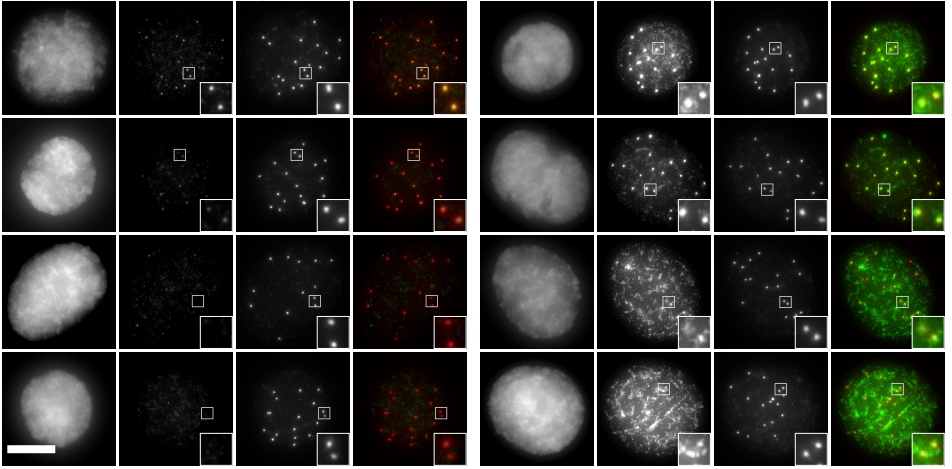
CDK phosphorylation of Xenopus laevis M18BP1 promotes its metaphase centromere localization
Bradley T French, Aaron F Straight
14-3-3 recruits keratin intermediate filaments to mechanically sensitive cell-cell contacts
Richard A. Mariani, Shalaka Paranjpe, Radek Dobrowolski, Gregory F. Weber
Gene editing can generate fragile bivalents in mouse oocytes
Marion Manil-Ségalen, Małgorzata Łuksza, Joanne Kannaan, Véronique Marthiens, Simon I.R. Lane, Keith T Jones, Marie-Émilie Terret, Renata Basto, Marie-Hélène Verlhac
Microtubule plus-tips act as signaling hubs for positioning the cleavage furrow during cytokinesis.
Vikash Verma, Thomas J Maresca
Systematic Characterization of RhoGEF/RhoGAP Regulatory Proteins Reveals Organization Principles of Rho GTPase Signaling
Paul Markus Mueller, Juliane Rademacher, Richard D Bagshaw, Keziban Merve Alp, Girolamo Giudice, Loise E Heinrich, Carolin Barth, Rebecca L Eccles, Marta Sanchez-Castro, Lennart Brandenburg, Geraldine Mbamalu, Monika Tucholska, Lisa Spatt, Celina Wortmann, Maciej T Czajkowski, Robert William Welke, Sunqu Zhang, Vivian Nguyen, Trendelina Rrustemi, Philipp Trnka, Kiara Freitag, Brett Larsen, Oliver Popp, Philipp Mertins, Chris Bakal, Anne-Claude Gingras, Olivier Pertz, Frederick P Roth, Karen Colwill, Tony Pawson, Evangelia Petsalaki, Oliver Rocks
Stick-slip dynamics of cell adhesion triggers spontaneous symmetry breaking and directional migration
Katharina Hennig, Irene Wang, Philippe Moreau, Leo Valon, Simon De Beco, Mathieu Coppey, Yekaterina Miroshnikova, Corinne Albiges Rizo, Cyril Favard, Raphael Voituriez, martial Balland
Excitable dynamics of Ras triggers self-organized PIP3 signaling for spontaneous cell migration
Seiya Fukushima, Satomi Matsuoka, Masahiro Ueda
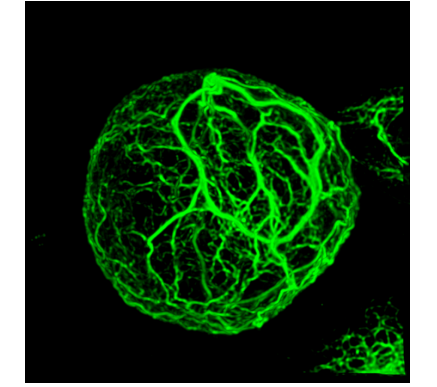
Vimentin filaments interact with the mitotic cortex allowing normal cell division
Sofia Duarte, Álvaro Viedma-Poyatos, Elena Navarro-Carrasco, Alma E Martínez, María A Pajares, Dolores Pérez-Sala
Force dependence of filopodia adhesion: involvement of myosin II and formins
Naila O. Alieva, Artem K. Efremov, Shiqiong Hu, Dongmyung Oh, Zhongwen Chen, Meenubharathi Natarajan, Hui Ting Ong, Antoine Jegou, Guillaume Romet-Lemonne, Jay T. Groves, Michael P. Sheetz, Jie Yan, Alexander D. Bershadsky
A combinatorial extracellular code tunes the intracellular signaling network activity to distinct cellular responses
Dmitry Kuchenov, Frederik Ziebell, Florian Salopiata, Mevlut Citir, Ursula Klingmueller, Wolfgang Huber, Carsten Schultz
Modelling
Turing patterns are common but not robust
Natalie S. Scholes, David Schnoerr, Mark Isalan, Michael P. H. Stumpf
Turing-like patterns can arise from purely bioelectric mechanisms
Micah Brodsky
Isolating and Quantifying the Role of Developmental Noise in Generating Phenotypic Variation
Maria A Kiskowski, Tilmann Glimm, Nickolas Moreno, Tony Gamble, Ylenia Chiari
Linear stability analysis of morphodynamics during tissue regeneration in plants
Anne-Mieke Reijne, Gunnar Pruessner, Giovanni Sena
Simulation of morphogen and tissue dynamics
M. D. Peters, L. D. Wittwer, A. Stopka, D. Barac, C. Lang, D. Iber
Stochastic and deterministic modelling of cell migration
Enrico Gavagnin, Christian A. Yates
A simplified multiphase multiscale model for tissue growth
E.C. Holden, B.S. Brook, S.J. Chapman, R.D. O’Dea
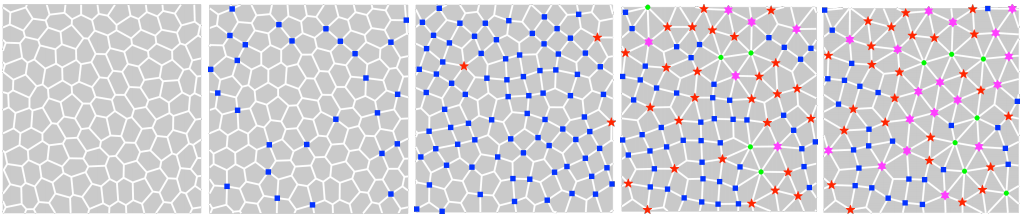
Rosette-driven rigidity transition in epithelial tissues
Le Yan, Dapeng Bi
Buckling without bending: a new paradigm in morphogenesis
T. A. Engstrom, Teng Zhang, A. K. Lawton, A. L. Joyner, J. M. Schwarz
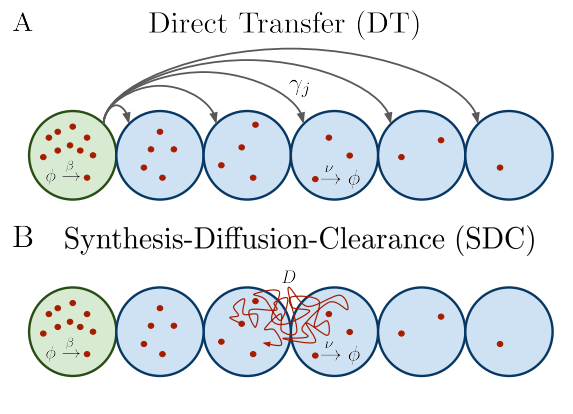
Diffusion vs. direct transport in the precision of morphogen readout
Sean Fancher, Andrew Mugler
Gradient Sensing via Cell Communication
Dallas Foster, Collin Victor, Brian Frost, Juan M. Restrepo
Modeling large fluctuations of thousands of clones during hematopoiesis: the role of stem cell self-renewal and bursty progenitor dynamics in rhesus macaque
Song Xu, Sanggu Kim, Irvin S. Y. Chen, Tom Chou
A minimal “push-pull” bistability model explains oscillations between quiescent and proliferative cell states.
Sandeep Krishna, Sunil Laxman
A Statistical Model of Cell Wall Dynamics during Expansive Growth
Shankar Lalitha Sridhar, Joseph K.E. Ortega, Franck Vernerey
Tools & resources
| Imaging etc.
BigStitcher: Reconstructing high-resolution image datasets of cleared and expanded samples
David Hörl, Fabio Rojas Rusak, Friedrich Preusser, Paul Tillberg, Nadine Randel, Raghav K. Chhetri, Albert Cardona, Philipp J. Keller, Hartmann Harz, Heinrich Leonhardt, Mathias Treier, Stephan Preibisch
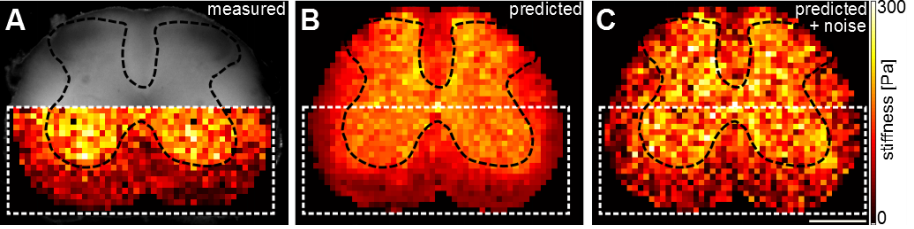
Predicting local tissue mechanics using immunohistochemistry
David E Koser, Emad Moeendarbary, Stefanie Kuerten, Kristian Franze
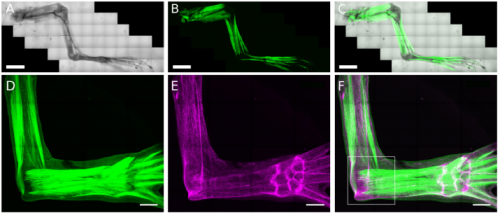
Broad applicability of a streamlined Ethyl Cinnamate-based clearing procedure
Wouter Masselink, Daniel Reumann, Prayag Murawala, Pawel Pasierbek, Yuka Taniguchi, Juergen A Knoblich, Elly M Tanaka
Single-embryo and single-blastomere immunoblotting reports protein expression heterogeneity in early-stage preimplantation embryos
Elisabet Rosas, Andrew J Modzelewski, Lin He, Amy E Herr
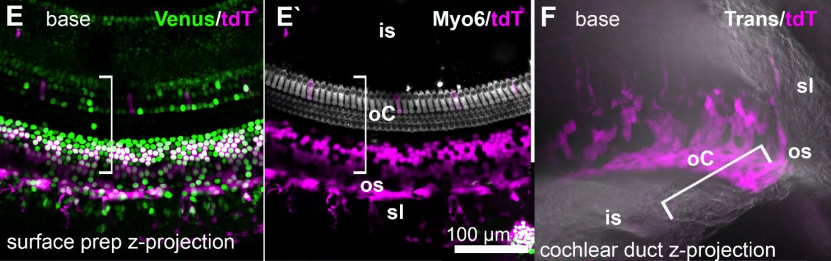
Fbxo2VHC mouse and embryonic stem cell reporter lines delineate in vitro-generated inner ear sensory epithelia cells and enable otic lineage selection and Cre-recombination
Byron H Hartman, Robert Böscke, Daniel C. Ellwanger, Sawa Keymeulen, Mirko Scheibinger, Stefan Heller
Collagen assembly and turnover imaged with a CRISPR-Cas9 engineered Dendra2 tag
Adam Pickard, Antony Adamson, Yinhui Lu, Joan Chang, Richa Garva, Nigel Hodson, Karl Kadler
Isolating mitotic and meiotic germ cells from male mice by developmental synchronization, staging, and sorting
Katherine A Romer, Dirk G de Rooij, David C Page
Generation of a versatile BiFC ORFeome library for analyzing protein-protein interactions in live Drosophila
Samir Merabet, Johannes Bischof, Marilyne Duffraisse, Edy Furger, Leiore Ajuria, Guillaume Giraud, Solene Vanderperre, Rachel Paul, Mikael Björklund, Damien Ahr, Alexis Ahmed, Lionel Spinelli, Christine Brun, Konrad Basler
DARPins recognizing mTFP1 as novel reagents for in vitro and in vivo protein manipulations
M. Alessandra Vigano, Dimitri Bieli, Jonas V Schaefer, Roman P Jakob, Shinya Matsuda, Timm Maier, Andreas Plueckthun, Markus Affolter
CDeep3M – Plug-and-Play cloud based deep learning for image segmentation of light, electron and X-ray microscopy
Matthias Haberl, Christopher Churas, Lucas Tindall, Daniela Boassa, Sebastien Phan, Eric A Bushong, Matthew Madany, Raffi Akay, Thomas J Deerinck, Steven T Peltier, Mark H Ellisman
Rapid micro fluorescence in situ hybridization in tissue sections
Deborah Huber, Govind V. Kaigala
Evaluation of Deep Learning Strategies for Nucleus Segmentation in Fluorescence Images
Juan C. Caicedo, Jonathan Roth, Allen Goodman, Tim Becker, Kyle W Karhohs, Claire McQuin, Shantanu Singh, Fabian Theis, Anne E. Carpenter
An Organometallic Ir(III) Molecular Probe for Imaging Microtubules in Fluorescence and Electron Microscopy
Xiaohe Tian Battaglia, Cesare De Pace, Lorena Ruiz-Perez, Bo Chen, Rina Su, Mingzhu Zhang, Ruilong Zhang, Qiong Zhang, Qin Wang, Hongping Zhou, Jieying Wu, Giuseppe Battaglia, Zhongping Zhang, Yupeng Tian
Kilohertz frame-rate two-photon tomography
Abbas Kazemipour, Ondrej Novak, Daniel Flickinger, Jonathan S Marvin, Jonathan King, Philip Borden, Shaul Druckmann, Karel Svoboda, Loren L Looger, Kaspar Podgorski
Embryo timelapses can be compiled and quantified to understand canonical histone dynamics across multiple cell cycles.
Lydia Smith, Paul S. Maddox
Convolutional Neural Network-Based Instance Segmentation Algorithm to Acquire Quantitative Criteria of Early Mouse Development
Yuta Tokuoka, Takahiro G Yamada, Noriko Hiroi, Tetsuya J Kobayashi, Kazuo Yamagata, Akira Funahashi
FluoEM: Virtual labeling of axons in 3-dimensional electron microscopy data for long-range connectomics
Florian Drawitsch, Ali Karimi, Kevin M Boergens, Moritz Helmstaedter
DataPackageR: Reproducible data preprocessing, standardization and sharing using R/Bioconductor for collaborative data analysis.
Greg Finak, Bryan Mayer, William Fulp, Paul Obrecht, Alicia Sato, Eva Chung, Drienna Holman, Raphael Gottardo
Forget Pixels: Adaptive Particle Representation of Fluorescence Microscopy Images
Bevan L. Cheeseman, Ulrik Günther, Mateusz Susik, Krzysztof Gonciarz, Ivo F. Sbalzarini
| Genome tools
Efficient single-copy HDR by 5′ modified long dsDNA donors
Jose Arturo Gutierrez-Triana, Thomas Thumberger, Tinatini Tavhelidse, Isabelle Thomas, Beate Wittbrodt, Tanja Kellner, Erika Tsingos, Joachim Wittbrodt
Switchable genome editing via genetic code expansion
Toru Suzuki, Maki Asami, Sanjay G. Patel, Louis Y. P. Luk, Yu-Hsuan Tsai, Anthony C. F. Perry
Spatiotemporally controlled genetic perturbation for efficient large-scale studies of cell non-autonomous effects
Rita Sousa-Nunes, Andrea Chai, Ana M Mateus, Fazal Oozeer
A Drosophila CRISPR/Cas9 toolkit for conditionally manipulating gene expression in the prothoracic gland as a test case for polytene tissues
Kirst King-Jones, Nhan Huynh
An expanded toolkit for gene tagging based on MiMIC and scarless CRISPR tagging in Drosophila
David Li-Kroeger, Oguz Kanca, Pei-Tseng Lee, Sierra Cowan, Michael Lee, Manish Jaiswal, Jose Louis Salazar, Yuchun He, Hugo Bellen
Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in C. elegans
Craig Mello, Gregoriy A Dokshin, Krishna S Ghanta, Katherine M Piscopo
An in vivo translation-reporter system for the study of protein synthesis in zebrafish embryos
Ines Palha, Isabelle Anselme, Sylvie Schneider-Maunoury, Francois Giudicelli
Enhancement of Transgene Expression by NF-Y and CTCF
Devon Zimmerman, Krupa Patel, Matthew Hall, Jacob Elmer
Precise tuning of gene expression output levels in mammalian cells
Yale S. Michaels, Mike B Barnkob, Hector Barbosa, Toni A Baeumler, Mary K Thompson, Violaine Andre, Huw Colin-York, Marco Fritzsche, Uzi Gileadi, Hilary M Sheppard, David JHF Knapp, Thomas A Milne, Vincenzo Cerundolo, Tudor A Fulga
Dynamics and spatial genomics of the nascent transcriptome in single mESCs by intron seqFISH
Sheel Shah, Yodai Takei, Wen Zhou, Eric Lubeck, Jina Yun, Noushin Koulena, Eric J. Liaw, Mina Amin, Long Cai
Branching developmental pathways through high dimensional single cell analysis in trajectory space
Denis Dermadi, Michael Bscheider, Kristina Bjegovic, Nicole H Lazarus, Agata Szade, Husein Hadeiba, Eugene C Butcher
SmartCodes : functionalized barcodes that enable targeted retrieval of clonal lineages from a heterogeneous population
Clare Rebbeck, Florian Raths, Bassem Ben Cheikh, Kenneth Gouin, Gregory Hannon, Simon Knott
Mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation
Bogdan Budnik, Ezra Levy, Guillaume Harmange, Nikolai Slavov
CRISPR-SURF: Discovering regulatory elements by deconvolution of CRISPR tiling screen data
Jonathan Y Hsu, Charles P Fulco, Mitchel A Cole, Matthew C Canver, Danilo Pellin, Falak Sher, Rick Farouni, Kendell Clement, James A Guo, Luca Biasco, Stuart H Orkin, Jesse M Engreitz, Eric S Lander, J. Keith Joung, Daniel E Bauer, Luca Pinello
Scarless gene tagging of transcriptionally silent genes in hiPSCs to visualize cardiomyocyte sarcomeres in live cells
Brock Roberts, Joy Arakaki, Kaytlyn A Gerbin, Haseeb Malik, Angelique Nelson, Melissa C Hendershott, Caroline Hookway, Susan A Ludmann, Irina A Mueller, Ruian Yang, Susanne M Rafelski, Ruwanthi N Gunawardane
A novel PCR method directly quantifies sequence features that block primer extension
Richard M Cawthon
SCuPhr: A Probabilistic Framework for Cell Lineage Tree Reconstruction
Hazal Koptagel, Seong-Hwan Jun, Jens Lagergren
Research practice & education
Researchers preferentially collaborate with same-gendered colleagues across the life sciences
Luke Holman, Claire Morandin
Career Choice, Gender, and Mentor Impact: Results of the U.S. National Postdoc Survey
Sean C. McConnell, Erica L. Westerman, Joseph F. Pierre, Erin J Heckler, Nancy B. Schwartz
Arbitration is needed to resolve scientific authorship disputes
Zen Faulkes
Becoming more transparent: Collecting and presenting data on biomedical Ph.D. alumni
Christopher L Pickett, Shirley Tilghman
Analysis and Correction of Inappropriate Image Duplication: The Molecular and Cellular Biology Experience
Arturo Casadevall, Elisabeth M Bik, Ferric C Fang, Amy Kullas, Roger J Davis
Reproducible big data science: A case study in continuous FAIRness
Ravi K Madduri, Kyle Chard, Mike D’Arcy, Segun C Jung, Alexis Rodriguez, Dinanath Sulakhe, Eric W Deutsch, Cory Funk, Ben Heavner, Matthew Richards, Paul Shannon, Gustavo Glusman, Nathan Price, Carl Kesselman, Ian Foster
Why not…
A skin pigment compound that also functions as sunglasses for birds
Malgorzata Zawadzka, Beatrix Racz, Dario Ambrosini, Carl Henrik Gorbitz, Jens Preben Morth, Elsa Lundanes, Frode Rise, Amund Ringvold, Steven Ray Wilson
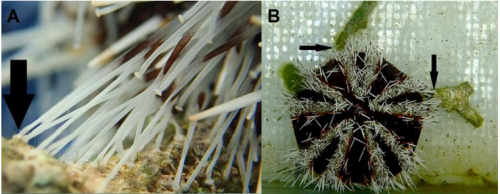
Tool use by four species of Indo-Pacific sea urchins
Glyn Barrett, Dominic Revell, Lucy Harding, Ian Mills, Axelle Jorcin, Klaus M. Stiefel
No support for the sexy-sperm hypothesis in the seed beetle: sons of monandrous females fare better in post-copulatory competition
Kristin A. Hook


 (2 votes)
(2 votes)