Nano Injections, Big Discoveries: The Journey to Map Neural and Inner Ear Lineages
Posted by Sandra de Haan, on 8 May 2025
Co-authored by Sandra de Haan and Jingyan He
In our recently published paper ‘Ectoderm barcoding reveals neural and cochlear compartmentalization‘, we utilized ultrasound-guided in utero nano injections to deliver heritable DNA barcodes to cells exposed to the amniotic fluid, performing the first high-throughput single cell lineage tracing study of the developing nervous system and inner ear. Our results led to the reclassification of cell lineages in the cochlea and provided a comprehensive single-cell atlas of neural and cochlear clonal relationships.
Sandra’s perspective: Work related to this publication already started a long time before I (Sandra) joined Emma R. Andersson’s lab back in 2019 to pursue my PhD studies. Work done by Katrin Mangold and Jingyan He, PhD students in the lab at the time, laid the foundation for successful in utero injections into the amniotic cavity to target the neural plate (Mangold et al, 2021). Like the neural plate, the progenitors of the inner ear are exposed to the amniotic fluid during an early time window of development (~E7 – E9) and we therefore hypothesized that we would be able to target the otic placode and manipulate progenitors of the inner ear using this technique. In fact, preliminary data from a Master’s student, Sanne Stokman, showed some targeting of the vestibular system of the inner ear. This data hinted that we should be able to target the cochlea as well – and laid the foundation to further explore this during my PhD studies. Drawing on expertise from two laboratories—the Andersson lab at the Karolinska Institute, specializing in developmental biology, Notch signaling, and in utero injection techniques, and Matt Kelley’s lab at the National Institutes of Health, experts in inner ear developmental biology—the first experiments targeting the cochlea were conducted in 2019. I vividly recall witnessing the targeting of the cochlea, for the first time, using low-titer H2B-GFP lentivirus injections performed by Jingyan. The mosaic-like targeting of hair cells along the cochlear spiral was truly remarkable (Fig. 1). I remember sharing these initial positive results with Emma via text while at the confocal microscope. Together with Jingyan, who truly mastered the injection technique, and together with the Infinigene core facility (established by Emma), we optimized our injection strategy to target the inner ear, including the injection volume, embryonic stage and viral titer. This journey was, of course, not without its challenges. At times, we faced difficulties with mouse breeding, low viral titers, and, not to forget, a pandemic that occurred along the way. Finally, in 2022, injections using high-titer virus yielded high-targeting efficiencies – of over 90% of the hair cells and supporting cells in the Organ of Corti. I remember observing the high efficiency targeting, quantifying targeting efficiencies late at night the same day, and presenting the results the next day over Zoom during the Kelley lab meeting – excited to share the new results.
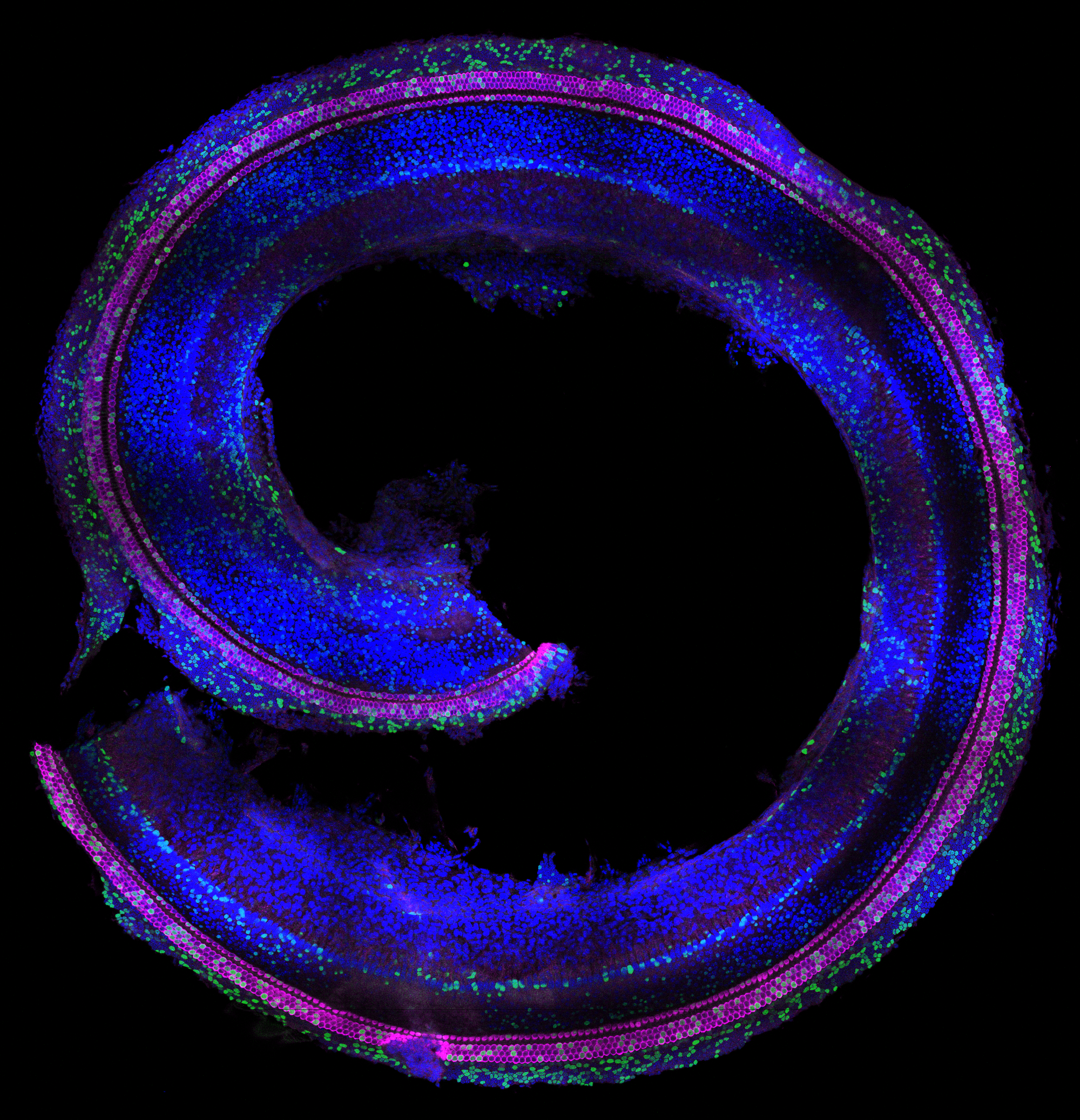
Using viral delivery of heritable DNA barcodes, we were able to perform high-throughput lineage trace studies in the inner ear for the first time, answering fundamental questions about inner ear development. I am still thrilled that we could apply this advanced method to explore lineage relationships within the inner ear. Although we initially focused on the divergence of medial and lateral cochlear cells, we soon realized that the most intriguing findings came from cells outside the Organ of Corti, including Hensen’s cells and populations within the stria vascularis. One of my favorite insights is the classification of Hensen’s cells. At the time, more research was performed on this specific population of cells in the cochlea, but no consensus existed on whether this cell type should be considered a supporting cell subtype or grouped with cells lateral to the Organ of Corti. Our data indicated that Hensen’s cells should be classified as lateral to the Organ of Corti, rather than being a supporting cell subtype of the Organ of Corti – if basing this definition on lineages. This classification might be relevant for future strategies focusing on regeneration and differentiation of cells within the cochlea. These findings also nicely aligned with results from another project of my PhD studies, in which we showed that Hensen’s cells respond differently to the loss of Jag1-mediated Notch activation compared to lateral supporting cells (De Haan et al 2024, Development).
As dissociation of cochlear cells relies on physical dislocation through microdissections, we inadvertently included cell types in our analysis that were not initially the focus of the study, including spiral ganglion neurons and glia. The analysis of barcode sharing between these populations proved to be quite complex. Contamination between spiral ganglion neurons and glia cells often occurs in single-cell preparations, so we investigated the barcode sharing between these populations to determine whether it was due to contamination or if subtypes of neurons might share a common origin with glia cells, which would challenge the current view. Ultimately, this experience taught me the importance of remaining open to new discoveries while ensuring that data collection and experimental design are well-suited to address the research questions. It highlighted the need for careful experimental planning, robust data collection and validation to draw accurate conclusions from complex datasets.
Jingyan’s perspective: While contributing to the inner ear lineage tracing project, I (Jingyan) was also focused on the overarching goal of my PhD projects: advancing the in utero injection technique to label non-ectodermal cells, building upon previous success in ectoderm targeting. By exploring different injection approaches, I successfully established a technique to label diverse cell types with other embryonic origins (ongoing work). As part of this highly ambitious project, we lineage traced tall ectoderm-derived cells using amniotic cavity injections at E7.5. We collected whole embryos at E9.5 and E10.5 after barcode labeling at E7.5 with amniotic cavity injection. This approach allowed us to study the lineage relations of central nervous system, neural crest-derived cells, as well as various epithelial lineages, including the otic lineage. This part of data was later incorporated into the inner ear lineage tracing paper to add a more comprehensive understanding of neurodevelopment and illustrate the potential clonal relations between otic epithelial lineages and other cell types.
One of the key challenges in this work was balancing the viral transduction efficiencies across different collection time points to ensure an optimal number of labeled cells for the single cell RNA sequencing and clonal analyses. Although E9.5 and E10.5 are just one day apart, the difference in total cell number is substantial. We had to pool a few E9.5 embryos to obtain sufficient cell numbers for clonal analyses. However, for E10.5, if we used the same amount of viral particle and reached the same transduction efficiency as E9.5 collection, a single E10.5 embryo yielded so many labeled cells that we needed to split them into multiple reactions when preparing the sequencing libraries.
Viral transduction efficiency itself was influenced by a variety of factors, such as the subtle differences of the embryos’ stages when injecting, viral storage time, freeze-thaw cycles and the variability between different virus production batches, making it difficult to consistently control the number of transduced cells recovered from each injection.
Collection days were always highly intense and stressful, involving a full workflow from the setting up of the cell sorter, embryo collection, dissection, dissociation, cell sorting, to library preparation, all within a tight window to preserve cell viability and RNA quality. Sandra and I always teamed up to streamline the workflow, assisting each other with reagent preparation, cell counting, and other time-sensitive steps.
Joint perspective: The manuscript, initially focused on cochlea only, was submitted shortly before Sandra’s successful PhD defense in August 2024. The E9.5/E10.5 ectodermal lineage tracing data was initially intended for a separate publication, but in response to reviewer and editorial feedback, we decided to incorporate it into the current paper during the revision process. This addition significantly enriched the manuscript and provided a more comprehensive understanding of neurodevelopment. As a result, Jingyan and Sandra shared the first authorship of the paper, highlighting the collaborative nature of our research and the importance of these findings.
Now that the paper has been published and new projects are underway in the Andersson lab applying this technique to different tissue systems, we are both excited to see how the technology will be used, further developed, and what biological insights it will uncover. We’re proud that our work also laid the foundation for an ERC Consolidator Grant to the Andersson lab, who will continue to push the frontiers of the technology – and the lab is looking for post docs!


 (1 votes)
(1 votes)