New 3D anatomy viewer available at eMouseAtlas
Posted by chris.armit@igmm.ed.ac.uk, on 26 January 2016
A new 3D viewer that allows interactive visualisation of mouse embryo anatomy is now available from the eMouseAtlas website (www.emouseatlas.org/). A slice viewer allows visualisation of anatomy on arbitrary section through mouse embryos – much like a virtual microtome – whilst a 3D anatomy pop-up window allows users to visualise the delineated anatomical components in an interactive 3D-context as either a wireframe or surface-rendered model. There is the additional option to change colour for selected anatomical components in both the slice viewer and the 3D anatomy pop-up window.
The new viewer uses IIP3D and WebGL technology to allow interactive exploration of 3D anatomy in a HTML5-compatible and WebGL-enabled web-browser and without the need for data download.
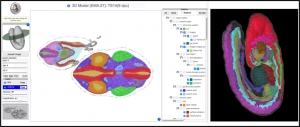
eMouseAtlas continues to develop tools and resources that enable high-end visualisation of embryo data. The anatomy is delineated to a very high standard and can be used for both research and for learning. There are future plans to explore use of this 3D viewer in web-based visualisation of 3D gene expression and phenotype data.


 (2 votes)
(2 votes)
Direct link to the model shown: http://www.emouseatlas.org/eAtlasViewer_ema/application/ema/anatomy/EMA27.php – click on the “3d view” button at the top right-hand side.