Out on a Flimb or: How I Learned to Stop Worrying and Trust the Mapping
Posted by mbrenthawkins, on 20 April 2021
Recently we reported the unexpected ability of fish mutants to develop limb-like bones in their pectoral fins (Hawkins et al., 2021). However, the most critical element of the study—finding these mutants in the first place—receives relatively little attention in the paper. Here I describe our efforts to find these monsters lurking inside the unassuming zebrafish.
The nuts and bolts of fins and limbs
The transition from fins to limbs is a defining transformation in vertebrate history and has served as a pivotal study system for comparative anatomy, paleontology, biomechanics, and developmental biology (Clack, 2009). Insight from each of these fields has illuminated different facets of how a relatively simple ancestral fin evolved into the complex arms and legs of tetrapods. Modern fins and limbs look quite different from one another but revealed in the fossil record are intermediate forms that connect their disparate morphologies (Jessen, 1972; Shubin et al., 2006; Zhu and Yu, 2009). By comparing the gene programs active in fins and limbs, we can ask which patterning mechanisms are common to both appendages, which mechanisms are derived in each, and which could be responsible for the changes in form found in evolution.
Tetrapod limbs have many long bones that articulate end on end away from the shoulder with distinct regions such as the upper arm (humerus), forearm (radius and ulna), and hand (Figure 1). In contrast, the pectoral fins of teleost fishes have just four long bones set side by side (proximal radials) followed distally by some small nodular bones (distal radials) and the dermal fin rays (Arratia, 1999). Non-teleost ray-finned fishes such as the bowfin have a slightly more impressive endoskeleton with additional articulations in the posterior part of the fin. Paleontological evidence suggests that the common ancestor of ray-finned fishes and tetrapods had a pectoral configuration much like that of the bowfin (Jessen, 1972, Zhu and Yu, 2009). In this scenario, teleosts such as the zebrafish represent a reduction of the ancestral appendage skeleton, while limbs exhibit its impressive elaboration (Coates, 1995).
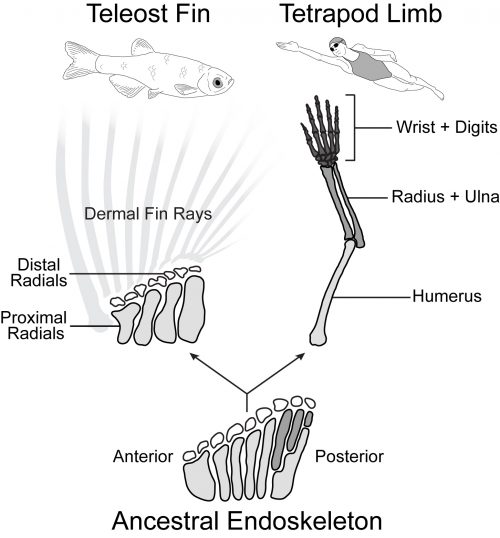
Forming an impressive body of work spanning the last four decades, developmental geneticists across the globe have discovered and characterized the manifold genes and pathways that control the growth and patterning in the nascent limb, leading to a deep understanding of the signaling ligands, receptors, and transcription factors necessary to make a normal appendage (Capdevila and Izpisúa Belmonte, 2001). Surprisingly, despite the morphological differences, many of these key limb patterning pathways are also expressed in developing teleost fins and play analogous (or conserved) functional roles (Mercader, 2007). There are differences in the expression and signaling function of some of these important players, but on the whole fin buds and limb buds behave quite like one another, and there is not one clear genetic factor present in limbs and absent in fins that is sufficient to imbue ‘limb-ness.’
Fishing for fin mutants
While assessing the role of candidate limb genes in growing fins has yielded critical insights into fin development, my colleagues Katrin Henke, Matthew Harris, and I decided to investigate the genes that can modify the zebrafish fin pattern using forward genetics. In a forward genetics approach, mutations are made at random and the investigator screens through mutated animals to pick out individuals with an interesting phenotype (Patton and Zon, 2001). Once an interesting mutant is found, we then work backwards using genetic mapping to determine which gene was mutated to cause the phenotype. The beauty of forward screens is that they let the organism tell you which genes are important to the process you want to know about. Sometimes you find an allele of a known essential regulator that has been extensively characterized, other times you find a gene that hasn’t been studied at all.
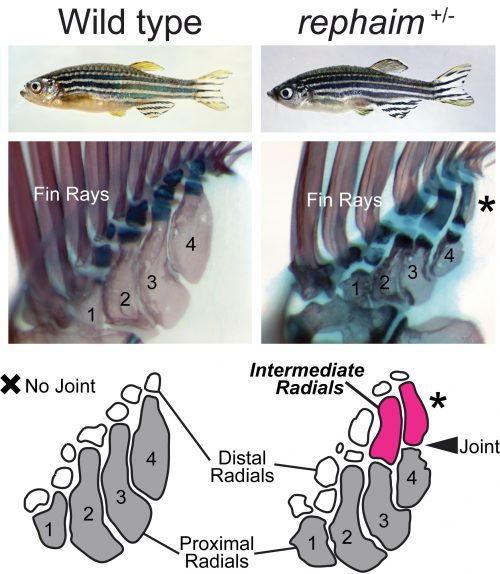
Katrin conducted several prodigious screens with a focus on mutants that affect the formation of the adult skeleton, and isolated hundreds of novel mutants (Henke et al., 2017). One mutant she picked out because of its modified pigmentation and dysmorphic fin rays was of particular interest to me and Matthew. Upon observing the internal skeleton under the dissecting scope, Matthew saw that the fin endoskeleton was affected and suggested I take a look. Shockingly, instead of having just the four long bones set side by side, this mutant had additional long bones forming in the distal endoskeleton that articulated with the proximal elements with a joint analogous to an elbow (Figure 2). This mutant didn’t just have fins, but what it had were not quite limbs either: it grew “flimbs.” At the time my dissertation project was focused on craniofacial mutants, but after this discovery (and Katrin’s blessing) my focus shifted to this fascinating mutant with limb-like fins. All good mutants need a name, and my friend and Old Testament scholar Maria suggested rephaim, a race of biblical giants fabled to have extra digits on their hands and feet.
After receiving a name, mutants need to be mapped to determine which gene is affected. Genetic mapping of mutations used to be a long and involved process using chromosomal markers to track linkage and recombination. In early zebrafish screens, a mutant line would be crossed to a wild-type fish from a different genetic background, and PCR-based methods would be used to find variable genomic positions and track down regions of DNA that segregated with the mutant phenotype (Knapik et al., 1996). In the last 15 years, however, the advent of next-generation sequencing made it possible to sequence mutants and their wild-type siblings to quickly identify genomic regions that associate with the mutant phenotype. To map rephaim, we utilized a whole-exome mapping approach to determine genomic regions that likely contain the causative mutation (Bowen et al., 2012). The mapping data gave us two putative regions that could contain the rephaim mutation, one on chromosome 4 and one on chromosome 9. Chromosome 4 has a reputation as being a nightmare for mapping, replete with inversions and transposons, and the implicated interval didn’t contain any interesting limb patterning genes. I did not like chromosome 4. On the other hand, the interval on chromosome 9 contained the HoxD cluster, a battery of genes with critical roles in appendage patterning and particularly implicated in the differential patterning of fins and limbs (Sordino et al., 1995; Freitas et al., 2012; Woltering et al., 2014). Not only would a mutation in a HoxD gene fit my expectations of what could cause a phenotype like rephaim, it would make subsequent analysis of the mutant phenotype much easier and fit well within the existing fin-to-limb literature. I even thought I might finish my dissertation early.
This, however, was not to be. Linkage analysis definitively ruled out chromosome 9 and the HoxD cluster. The initial mapping signal that I had pinned my hopes on was due to a block of genetic homogeneity that was shared between mutants and wild-type siblings, meaning that both mutants and wild-type animals had the same alleles in this region and thus could not contain the causative mutation. Meanwhile, additional recombination analysis strengthened the association of rephaim with chromosome 4 and narrowed the linkage interval to a small window containing just one coding mutation in a gene called wiskott-aldrich syndrome protein like-b (waslb). Unlike my precious HoxD cluster, the waslb gene was not a known regulator of limb development, and everything I saw in the literature gave the impression that it was a “housekeeping” gene: ubiquitous expression, essential functions in actin metabolism, and involvement in myriad cellular pathways (Snapper and Rosen, 1999). Around this time, we were also mapping a second mutant with a similar phenotype, a fish called wanda (van Eeden et al., 1996; Haffter et al., 1996). The causative mutation for wanda mapped to the gene vav2, a similarly unexciting locus from a skeletal patterning perspective (Hornstein et al., 2004). I felt the path to my PhD lengthening in real time.
X marks the spot on a genetic treasure map
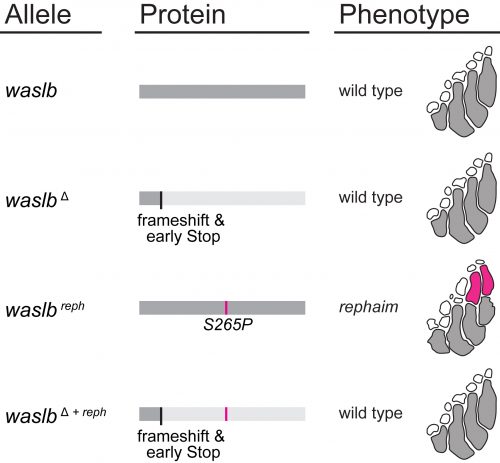
Nevertheless, all the mapping data pointed to waslb and vav2, so these genes demanded our attention. Mapping implicated these genes, but we still needed to experimentally confirm their role in the flimb phenotype. We used CRISPR-Cas9 to make loss-of-function alleles, but even homozygous null mutants had a wild-type phenotype (Figure 3). Next we tried injecting mutant mRNA into the embryo but saw no effect, likely due to the late appearance of the phenotype. In a final push to demonstrate the causative nature of the waslb and vav2 mutations, we used CRISPR to create frameshift lesions in cis to the candidate mutations and knockout the mutant alleles specifically. When the mutant copies of waslb and vav2 were removed, we rescued the phenotype and reverted the mutants to wild-type fin patterning. There was no doubt, mutations in waslb and vav2 cause the flimb phenotype. But this left us with a bigger question, how in the world are these genes changing skeletal patterning? I came up with an axiom to sooth myself: “if good science raises more questions than it answers, then the best science must raise only questions and answer nothing.”
However, the wealth of limb patterning knowledge established by developmental geneticists was able to guide our inquiry. As mentioned earlier, Hox genes have critical functions in the patterning and growth of limb bones along the proximal-distal axis, and recent studies had revealed that Hox13 was required for the formation of the most distal structures in fins just like in limbs (Nakamura et al., 2016). We thought that the new bones in rephaim might also share this distal Hox13 regionality, and crossed rephaim into a Hox13-null genetic background. To our surprise, we found that loss of Hox13 actually enhanced the flimb phenotype and resulted in the formation of even more bones along the distal aspect of the endoskeleton (Figure 4). Hoxa13 is known to negatively regulate Hoxa11 expression in the limb (Kherdjemil et al., 2016), and we thought the enhanced phenotype might be the result of derepression of Hox11 genes. Around this time we also were analyzing limb-specific Wasl knockout mice, and observed limb defects similar to those seen in Hoxa11 mouse mutants. Intriguingly, Hox11 genes are also required for the normal development for the bones in the middle region of the limb, the radius and ulna (Davis et al., 1995).
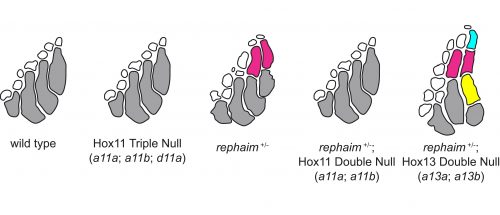
Following these clues, we generated null alleles of the hoxa11 and hoxd11 paralogs in the zebrafish. While loss of these genes had no effect on fin patterning in a wild-type background, we found that loss of hoxa11a and hoxa11b prevented the formation of the extra bones in rephaim mutants (Figure 4). Moreover, we generated knock-in hoxa11b reporter zebrafish and found that rephaim and wanda mutants cause the upregulation of hoxa11b expression. These results were quite interesting: even though the Hox11 paralogs are not required for normal fin patterning in zebrafish, they still possess the ability to specify the formation of an intermediate long bone position along the proximal-distal axis of the appendage skeleton. This suggests that the capacity to specify ‘middle’ and ‘distal’ regions is not unique to limbs, but was present in the common ancestor of ray- and lobe-finned fishes. Although not expressed in teleosts, this developmental potential has been retained in a latent state and can be redeployed by simple perturbations.
Wasl, Hox, and Beyond
This forces me to wonder what other latent limb patterning mechanisms that might reside in the developing fin bud, and how waslb is able to activate at least some of them. The mechanistic connection between waslb, vav2, and Hox regulation is an open question. In part, we know that waslb mediates the formation of F-actin foci that colocalize with Hox-positive cells in the distal fin (Hawkins et al., 2021). Given its roles in cell motility it could be that waslb effects the migration of these cells, but there are many other possibilities. Wasl also directly regulates transcription (Wu et al., 2006), modulates Wnt (Lyubimova et al., 2010) and TGFB (Lefever et al., 2010) signaling, and is directly involved in the colinear activation of the HoxB cluster (Ferrai et al., 2009). Then again, there could be another pathway that we do not yet understand. I will go out on a flimb here and say the zebrafish still has more to tell us about these mechanisms…as long as we are willing to trust the mapping.
For further reading, you can find our published manuscript at https://www.cell.com/cell/fulltext/S0092-8674(21)00003-9
You can also see an explainer thread on Twitter at https://twitter.com/Homeobox/status/1357382005261017098?s=20
References
Arratia, G. (1999). The monophyly of Teleostei and stem-group teleosts. Mesozoic Fishes, 2, 265–334.
Bowen, M. E., Henke, K., Siegfried, K. R., Warman, M. L., & Harris, M. P. (2012). Efficient mapping and cloning of mutations in zebrafish by low-coverage whole-genome sequencing. Genetics, 190(3), 1017–1024.
Capdevila, J., & Izpisúa Belmonte, J. C. (2001). Patterning mechanisms controlling vertebrate limb development. Annual Review of Cell and Developmental Biology, 17, 87–132.
Clack, J. A. (2009). The Fin to Limb Transition: New Data, Interpretations, and Hypotheses from Paleontology and Developmental Biology. Annual Review of Earth and Planetary Sciences, 37(1), 163–179.
Coates, M. I. (1995). Limb evolution. Fish fins or tetrapod limbs–a simple twist of fate? Current Biology: CB, 5(8), 844–848.
Davis, A. P., Witte, D. P., Hsieh-Li, H. M., Potter, S. S., & Capecchi, M. R. (1995). Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature, 375(6534), 791–795.
Ferrai, C., Naum-Onganía, G., Longobardi, E., Palazzolo, M., Disanza, A., Diaz, V. M., Crippa, M. P., Scita, G., & Blasi, F. (2009). Induction of HoxB transcription by retinoic acid requires actin polymerization. Molecular Biology of the Cell, 20(15), 3543–3551.
Freitas, R., Gómez-Marín, C., Wilson, J. M., Casares, F., & Gómez-Skarmeta, J. L. (2012). Hoxd13 contribution to the evolution of vertebrate appendages. Developmental Cell, 23(6), 1219–1229.
Haffter, P., Odenthal, J., Mullins, M. C., Lin, S., Farrell, M. J., Vogelsang, E., Haas, F., Brand, M., van Eeden, F. J., Furutani-Seiki, M., Granato, M., Hammerschmidt, M., Heisenberg, C. P., Jiang, Y. J., Kane, D. A., Kelsh, R. N., Hopkins, N., & Nüsslein-Volhard, C. (1996). Mutations affecting pigmentation and shape of the adult zebrafish. Development Genes and Evolution, 206(4), 260–276.
Hawkins, M. B., Henke, K., & Harris, M. P. (2021). Latent developmental potential to form limb-like skeletal structures in zebrafish. Cell. https://doi.org/10.1016/j.cell.2021.01.003
Henke, K., Daane, J. M., Hawkins, M. B., Dooley, C. M., Busch-Nentwich, E. M., Stemple, D. L., & Harris, M. P. (2017). Genetic Screen for Postembryonic Development in the Zebrafish (Danio rerio): Dominant Mutations Affecting Adult Form. Genetics, 207(2), 609–623.
Hornstein, I., Alcover, A., & Katzav, S. (2004). Vav proteins, masters of the world of cytoskeleton organization. Cellular Signalling, 16(1), 1–11.
Jessen, H. L. (1972). Fossils and Strata, Schultergurtel und Pectoralflosse bei Actinopterygiern. Lethaia, 5(3), 344.
Kherdjemil, Y., Lalonde, R. L., Sheth, R., Dumouchel, A., de Martino, G., Pineault, K. M., Wellik, D. M., Stadler, H. S., Akimenko, M.-A., & Kmita, M. (2016). Evolution of Hoxa11 regulation in vertebrates is linked to the pentadactyl state. Nature, 539(7627), 89–92.
Knapik, E. W., Goodman, A., Atkinson, O. S., Roberts, C. T., Shiozawa, M., Sim, C. U., Weksler-Zangen, S., Trolliet, M. R., Futrell, C., Innes, B. A., Koike, G., McLaughlin, M. G., Pierre, L., Simon, J. S., Vilallonga, E., Roy, M., Chiang, P. W., Fishman, M. C., Driever, W., & Jacob, H. J. (1996). A reference cross DNA panel for zebrafish (Danio rerio) anchored with simple sequence length polymorphisms. Development , 123, 451–460.
Lefever, T., Pedersen, E., Basse, A., Paus, R., Quondamatteo, F., Stanley, A. C., Langbein, L., Wu, X., Wehland, J., Lommel, S., & Brakebusch, C. (2010). N-WASP is a novel regulator of hair-follicle cycling that controls antiproliferative TGF{beta} pathways. Journal of Cell Science, 123(Pt 1), 128–140.
Lyubimova, A., Garber, J. J., Upadhyay, G., Sharov, A., Anastasoaie, F., Yajnik, V., Cotsarelis, G., Dotto, G. P., Botchkarev, V., & Snapper, S. B. (2010). Neural Wiskott-Aldrich syndrome protein modulates Wnt signaling and is required for hair follicle cycling in mice. The Journal of Clinical Investigation, 120(2), 446–456.
Mercader, N. (2007). Early steps of paired fin development in zebrafish compared with tetrapod limb development. Development, Growth & Differentiation, 49(6), 421–437.
Nakamura, T., Gehrke, A. R., Lemberg, J., Szymaszek, J., & Shubin, N. H. (2016). Digits and fin rays share common developmental histories. Nature, 537(7619), 225–228.
Patton, E. E., & Zon, L. I. (2001). The art and design of genetic screens: zebrafish. Nature Reviews. Genetics, 2(12), 956–966.
Shubin, N. H., Daeschler, E. B., & Jenkins, F. A., Jr. (2006). The pectoral fin of Tiktaalik roseae and the origin of the tetrapod limb. Nature, 440(7085), 764–771.
Snapper, S. B., & Rosen, F. S. (1999). The Wiskott-Aldrich syndrome protein (WASP): roles in signaling and cytoskeletal organization. Annual Review of Immunology, 17, 905–929.
Sordino, P., van der Hoeven, F., & Duboule, D. (1995). Hox gene expression in teleost fins and the origin of vertebrate digits. Nature, 375(6533), 678–681.
van Eeden, F. J., Granato, M., Schach, U., Brand, M., Furutani-Seiki, M., Haffter, P., Hammerschmidt, M., Heisenberg, C. P., Jiang, Y. J., Kane, D. A., Kelsh, R. N., Mullins, M. C., Odenthal, J., Warga, R. M., & Nüsslein-Volhard, C. (1996). Genetic analysis of fin formation in the zebrafish, Danio rerio. Development , 123, 255–262.
Woltering, J. M., Noordermeer, D., Leleu, M., & Duboule, D. (2014). Conservation and divergence of regulatory strategies at Hox Loci and the origin of tetrapod digits. PLoS Biology, 12(1), e1001773.
Wu, X., Yoo, Y., Okuhama, N. N., Tucker, P. W., Liu, G., & Guan, J.-L. (2006). Regulation of RNA-polymerase-II-dependent transcription by N-WASP and its nuclear-binding partners. Nature Cell Biology, 8(7), 756–763.
Zhu, M., & Yu, X. (2009). Stem sarcopterygians have primitive polybasal fin articulation. Biology Letters, 5(3), 372–375.


 (4 votes)
(4 votes)