Reshaping morphogen gradients, one miRNA at a time
Posted by Marcos Simoes-Costa, on 15 March 2021
Jacqueline Copeland and Marcos Simoes-Costa
Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY, USA
The neural crest has long been referred to as “the fourth germ layer” for its remarkable ability to give rise to a number of cell types in the vertebrate embryo including neurons, glia, bone, and cartilage. Specified at the border of the neural plate, the neural crest is intimately linked to this adjacent cell population, which will give rise to the central nervous system. Directing the partitioning of the ectoderm into these two territories are inductive interactions driven by signaling systems including Wnt, BMP, and FGF. These signals form overlapping spatial gradients across different ectodermal territories, driving the regulatory programs required to induce the neural crest and the neural plate. Many developmental biologists have studied the timing and levels of signaling required for proper neural crest formation, but fundamental questions remain: How do distinct combinations of signaling molecules give rise to different cell types? Furthermore, how do fields of cells interpret and respond to these signals?
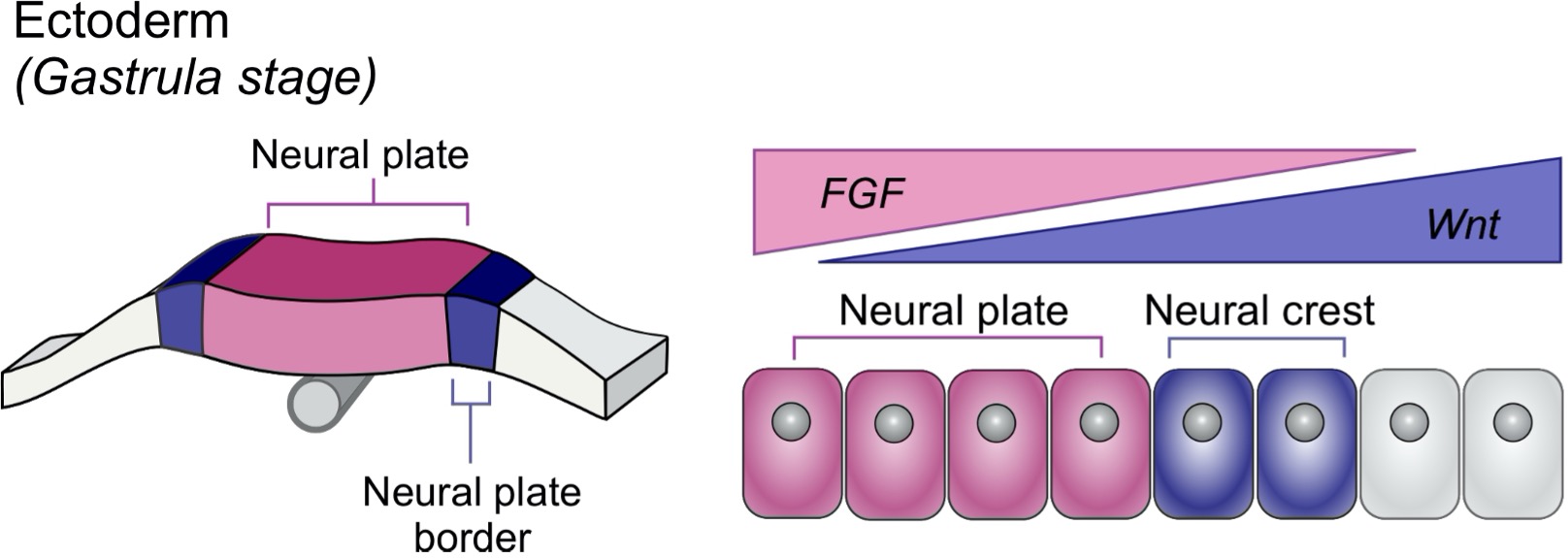
A conceptual framework for understanding these questions was established by Lewis Wolpert in 1969 with his seminal paper “Positional Information and the Spatial Pattern of Cellular Differentiation”1. Wolpert’s “French Flag Model” proposed that spatial gradients of signaling molecules known as morphogens drive subdivision of cellular fields based on concentration. Although Wolpert’s model offered foundational insights, today we know these processes are much more complex. For example, there must exist mechanisms by which cells can produce binary responses to distinct ranges of signaling levels. One such mechanism relies on the transcriptional control of signaling system activity through activation of agonists/antagonists. However, transcription can be a leaky process, and may not confer the optimal levels of signaling activity needed for cell fate decisions. Therefore, other modes of regulation may offer robustness in these developmental programs.
When I began my first year of graduate school at Cornell University I had never even heard of Lewis Wolpert or the neural crest, for that matter. I was first made aware of this cell population after hearing a rotation talk from my now mentor Marcos Simoes-Costa. Although I had no background in development, I was amazed by the beautiful movies of migrating neural crest cells and the chick model system’s tractability for investigating gene regulatory programs. After Marcos’s talk, I was very excited to rotate in his lab and eager to discuss rotation projects. Prior to my rotation, Marcos had published his first article as a new PI, investigating the role of the Lin28/Let-7 axis in neural crest multipotency2. From this work he became interested to look further at the post-transcriptional regulation of this cell type. As an undergraduate, I studied genetic interactions associated with pre-mRNA splicing in Saccharomyces Cerevisiae. Through this work, I developed an immense interest in post-transcriptional gene regulation, which I consider the underdog of the central dogma of molecular biology. As luck would have it, our interests aligned, and we embarked on a new project which culminated in a recent publication in PNAS3.
It has been known for roughly a decade that Dicer, a key enzyme in the miRNA biogenesis pathway, is critical for neural crest development and differentiation. Conditional Dicer knockout mice exhibit severe craniofacial defects due to lack of neural crest-derived structures, highlighting the importance of Dicer, and therefore mature miRNA species, in the formation of this cell type4-6. However, few studies had investigated the mechanistic roles that miRNAs and their gene targets may play in this cell type. This led us to our major goal of identifying and characterizing miRNA function during early neural crest formation.
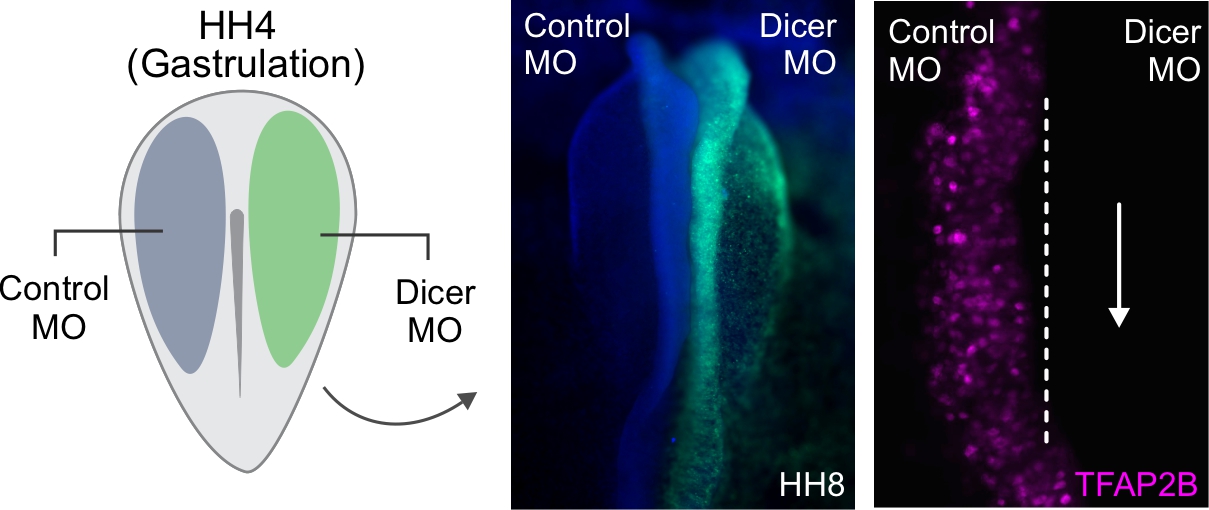
It would be quite difficult to assay the phenotypic outcome of inhibiting every individual miRNA expressed in the chicken genome. So, to get an idea of the role miRNAs might be playing during neural crest specification, we first turned to Dicer, examining its expression in relation to that of the neural crest specification marker TFAP2B. What came as quite a surprise was that Dicer, which is thought to be a ubiquitously expressed protein, was enriched in neural crest cells. Furthermore, through interrogating the Dicer locus, we uncovered a neural crest-specific enhancer driving Dicer upregulation in this cell population. This was an exciting finding, as it demonstrated that there may be increased processing and turnover of miRNAs within neural crest cells. This idea made a lot of sense to us, as neural crest cells undergo rapid genomic and morphological changes in their relatively short lifetime, which may be facilitated by different groups of miRNAs.
The chick embryo offers a beautiful system in which we can perform bilateral electroporations to observe control and knockdown phenotypes within the same organism. Using this method, we injected gastrulating chick embryos with a Dicer protein-inhibiting morpholino. Since the phenotypic effects of morpholino based knockdown can be quite broad, we performed a Nanostring analysis in order to more globally assess the effects of Dicer knockdown during neural crest specification. To our surprise, knockdown of Dicer not only resulted in a loss of neural crest markers but was also accompanied by increased expression of several neural plate-specific factors. Observing knockdown embryos, we indeed saw an expansion of the neural plate at the expense of neighboring neural crest cells.
These observations led us to hypothesize that miRNAs (synthesized via Dicer) may play an important role in the cell fate decision between the neural crest and neural plate. To test this hypothesis, we needed to isolate these cell populations in order to identify the abundant and unique miRNAs present in each. For this we turned to a commonly used system in our lab, in which we can inject chick embryos with cell type-specific enhancer reporters and then perform FACS to isolate pure populations of cells. Since we were interested in the neural plate and the neural crest, we isolated cells from each of these populations and performed small RNA-sequencing. Admittedly, getting small RNA-sequencing to work was a feat in itself. Most of my rotation and my first summer in lab were spent optimizing the protocol for small numbers of sorted chick neural crest cells. This involved dissecting hundreds of chick embryos but was accompanied by lots of time with Marcos at the electroporation station discussing what miRNAs could be doing in crest.
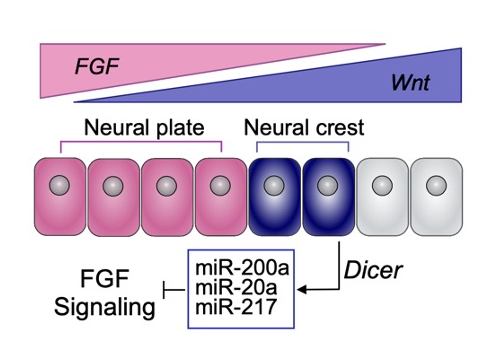
Once we finally optimized a protocol for small RNA-sequencing, we were off to the races, identifying miRNAs and determining their modes of action. Through several analyses including miRNA target prediction, we ultimately identified a group of neural crest miRNAs that target components of the FGF signaling pathway. This was an “aha!” moment for us as we knew levels of FGF must be quite precise for neural crest induction: too much and the cells are fated towards the neural plate, but not enough, and neural crest will not form at all. We hypothesized this group of miRNAs was required in crest to keep levels of FGF low, inhibiting a neural plate fate, and confirmed this through several functional approaches. One of my favorite experiments we performed, that truly wrapped up the paper, was the rescue of the Dicer knockdown phenotype by adding back our FGF-targeting neural crest miRNAs. Getting back to post-transcriptional regulation being the underdog of the central dogma, this was amazing to me. miRNAs typically have very modest repressive effects (less than 1.5 fold) on gene expression. Given this, I was not sure if by putting back our FGF-targeting miRNAs we would rescue the Dicer knockdown phenotype. But to my surprise, by working in a concerted fashion to target different FGF pathway components and attenuate levels of FGF, these three miRNAs were quite capable of rescuing the neuralization switch observed upon Dicer knockdown.
I am quite proud of this paper, not only because it is my first publication in graduate school, but because it tells a story of how small RNAs can work together to drastically influence the cell fate decisions of neighboring cell populations in the ectoderm. I also appreciate how it unifies two opposite ends of the of neural crest regulatory spectra: signaling system inputs that jumpstart the neural crest GRN and post-transcriptional regulation of these signaling systems to ensure the proper thresholds of activity are met. Moving forward, I would like to explore regulation of signaling systems via miRNAs at a broader level, considering their contribution to patterning events related to other tissue types, as well as the regulation of the miRNAs that reside in those tissues. I also hope that this work can inform upon other developmental scenarios where tuning of signaling systems via miRNAs is critical for cell fate commitment.
- Wolpert, L., Positional information and the spatial pattern of cellular differentiation. J Theor Biol, 1969. 25(1): p. 1-47.
- Bhattacharya, D., et al., Control of neural crest multipotency by Wnt signaling and the Lin28/let-7 axis. Elife, 2018. 7.
- Copeland, J. and M. Simoes-Costa, Post-transcriptional tuning of FGF signaling mediates neural crest induction. Proc Natl Acad Sci U S A, 2020. 117(52): p. 33305-33316.
- Zehir, A., et al., Dicer is required for survival of differentiating neural crest cells. Dev Biol, 2010. 340(2): p. 459-67.
- Huang, Z.P., et al., Loss of microRNAs in neural crest leads to cardiovascular syndromes resembling human congenital heart defects. Arterioscler Thromb Vasc Biol, 2010. 30(12): p. 2575-86.
- Huang, T., et al., Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J Mol Cell Biol, 2010. 2(3): p. 152-63.


 (3 votes)
(3 votes)