Revisiting the classics: coupling embryology with genomics to alter cell fate
Posted by Marcos Simoes-Costa, on 13 July 2016
Comment on “Reprogramming of avian neural crest axial identity and cell fate“, Science 352, 1570-1573, (2016).
Marcos Simoes-Costa, Department of Molecular Biology and Genetics, Cornell University
Marianne Bronner, Division of Biology and Biological Engineering, California Institute of Technology
In the 19th century, most embryologists (i.e. precursors to developmental biologists) accepted the germ layer theory – the idea that embryos are composed of distinct layers (ectoderm, endoderm and mesoderm), which will give rise to specific tissues and cell types in the adult. For instance, it was believed that all skeletal components of the body were derived from the mesoderm, whereas the ectoderm differentiated into the nervous system and epidermis. Around the turn of the 20th century, Julia Platt discovered a remarkable exception to this. She used salamander embryos to track neural crest cells, which are an ectodermal progenitor cell population, and observed that they gave rise to skeletal elements of the head (Platt, 1883). These findings were so controversial at the time that Platt was unable to get a faculty position, quit science, and became a politician. But a hundred and twenty years have passed since Platt’s results were published, and time has been kind to her legacy. Today, we know with certainty that the neural crest is indeed an exceptional cell type. During early formation of the central nervous system, neural crest cells delaminate from the neural tube, engage in extensive migration and give rise to a large portion of craniofacial skeleton as well as several other tissues.
This particular question – how ectodermal cells can differentiate into skeletal tissue– captured my imagination and drove my interest toward neural crest biology. Not that this was the only remarkable feature of neural crest cells: they have the ability to undergo epithelial to mesenchymal transition, are migratory, and can to differentiate into multiple cell types. While neural crest cells emerge from the central nervous system along the length of the body axis, only the cranial crest can give rise to the cartilage and bone of the face. This was elegantly demonstrated by Nicole Le Douarin in the 1960s and -70s through the use of chicken-quail chimeras (reviewed in Le Douarin, 1982). Le Douarin and colleagues showed that trunk neural crest cells are unable to form cartilage even if grafted to the head. Strikingly, cranial neural crest grafted to the trunk gave rise to ectopic cartilage nodules, indicating that these cells are autonomously capable of differentiating into mesenchymal tissue. These results demonstrated that the cranial neural crest was intrinsically different from the other crest subpopulations.
Joining Marianne Bronner’s lab at Caltech after finishing my Ph.D. in Brazil, I wanted to learn molecular biology and genomics. The lab is very interested in gene regulatory control of neural crest specification, so my first project, in collaboration with Tatjana Sauka-Spengler and Sonja Mckeown, focused on cis-regulatory control of the transcription factor FoxD3 in chick embryos. FoxD3 is a classic neural crest marker expressed by all neural crest cells along the entire body axis. However, we discovered different enhancers that drove FoxD3 expression in the head versus the trunk: a distal regulatory element (NC2) mediated trunk expression whereas a unique cranial-specific enhancer closer to the FoxD3 promoter (NC1) drove expression in the head. This suggested that axial differences were encoded in the genome (Simoes-Costa et al., 2012), bringing me right back to the question of what makes the cranial neural crest unique. Importantly, our identification of a axial-specific neural crest enhancers provided a new tool for exploring the molecular mechanisms that underlie the development of the cranial crest.
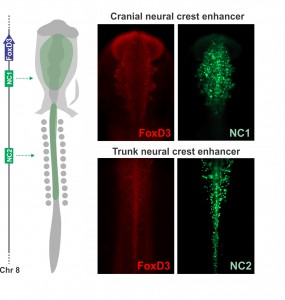
The strategy to investigate the molecular basis of the cranial neural crest’s ability to give rise to cartilage emerged gradually during daily meetings with Marianne, mostly at the coffee machine. During these informal conversations, we delineated an experimental approach to identify factors unique to the cranial crest that drove differentiation into chondrocytes. Marianne had been interested in this question for a long time, and for more personal reasons – while recovering from a bicycle accident (face plant resulting in a broken nose and deviated septum), she learned from the craniofacial surgeon that facial cartilage is very difficult to replace. We decided to employ a combination of comparative transcriptomics, gene regulatory analysis, and classical embryology to identify the molecular players that endow the cranial neural crest with its ability to form cartilage. Crucial to this approach was the fact that the trunk neural crest is unable to make cartilage in vivo. We predicted that by comparing the transcriptomes of the cranial to the trunk populations – that now could be labelled and isolated with the NC1 and NC2 enhancers – we would be able to identify the transcriptional circuitry that activates chondrocytic differentiation in cranial neural crest cells.
Previously, only a couple factors had been identified as being specific to the cranial crest. Id2 was found to be a cranial neural crest gene by a differential display screen performed by Brad Martinsen in Marianne’s lab almost twenty years ago (Martinsen and Bronner-Fraser, 1998). Eric Theveneau and Muriel Altabef also had identified Ets1 as an important player in the development of this cell population (Theveneau et al, 2007). Our comparative transcriptome analysis identified multiple transcription factors that are enriched in the cranial crest, and we proceeded to organize them in a cranial-specific neural crest regulatory circuit. By analyzing their expression patterns during early embryonic development, we defined the hierarchy of the circuit; and by methodically inactivating each gene, we identified the links that integrated these multiple components into a regulatory network.
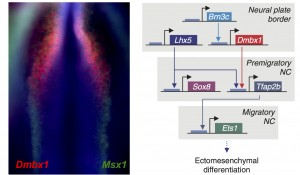
We were very excited by the identification of this novel cranial-specific regulatory circuit, as it provided important insights about the biology of the cranial neural crest. For instance, it showed that expression of cranial neural crest genes depends upon anterior regulatory information that is laid out during gastrulation. Brn3c, Lhx5, and Dmbx1, which are the components of the first “early” level of the circuit, are all co-expressed adjacent to the anterior neural plate in the early gastrula. These genes are required for the expression of the “later” genes – Sox8, Tfap2b and Ets1 – which are retained in the migratory cranial neural crest. However, one fundamental question remained – could this regulatory circuit be important for the cranial neural crest’s potential to give rise to cartilage and bone?
During our coffee break conversations, Marianne and I came to the conclusion that the best way to tackle this question would be to transplant components of the cranial circuit to trunk cells and see if they would gain the ability to switch fates and differentiate into skeletal tissue. Thus, we decided to revisit the classical experiments of Nicole Le Douarin, but with a twist – we would transfect the trunk neural crest from the donor embryos with components of the cranial circuit before grafting them to the head. We would also trade the quail embryos in favor of transgenic GFP+ chicken embryos, to facilitate tracking of the grafted cells. After surgery, we would let our chimeras grow for a week and verify if the reprogrammed trunk crest differentiated into facial skeleton. At that time, I was eager for a foray into classic cut and paste embryology, thinking that perhaps I would spend a month performing surgeries and a couple of weeks analyzing the chimeric embryos.
Despite my optimism, that was not the case. Grafting is an art and the chimeric embryos are unforgiving of shaky hands. In my daily meetings with Marianne, I was now drinking decaffeinated tea. Since the grafting experiments were taking an enormous amount of time, we realized we needed a more efficient strategy to identify which components of the cranial circuit were able to reprogram the trunk neural crest. And thus, we came up with the idea of using neural crest axial specific enhancers as reporters of cranial identity. The Sox10E2 enhancer from the Sox10 gene, which was identified by Paola Betancur in Marianne’s lab (Betancur et al., 2010), was perfect for this since it is active in migratory cranial neural crest cells and inactive in the trunk. Thus, we proceeded to transfect trunk cells with all different combinations of the cranial circuit components attempting to activate the cranial enhancer in the trunk neural crest.
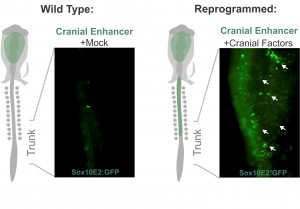
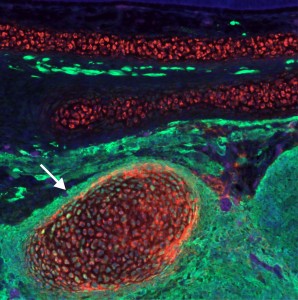
These experiments lead to the discovery that Sox8, Tfap2b and Ets1 are able turn on the cranial enhancer in the trunk and also to increase expression levels of endogenous cranial genes in these neural crest cells. When the reprogrammed trunk neural crest cells were grafted in the head of a host embryo, they not only gave rise to the normal trunk neural crest cell derivatives, melanocytes and neurons, but these grafts also differentiated into chondrocytes, forming conspicuous cartilage nodules. And thus we had demonstrated that we could use information decoded through gene regulatory studies to reprogram neural crest identity and change its fate in vivo.
I am thrilled with the publication of this paper not only because it represents a great deal of hard work, but also because it answers a long-standing question that has interested me for years. The aspect of the work that I like the most is the marriage between old and new – our results highlight the power of classical embryological approaches, especially when used in conjunction with modern techniques. Moving forward, we will employ these findings to devise novel strategies for in vitro differentiation of facial cartilage. It is our hope that regulatory information derived from embryonic development can inform upon novel protocols for cell and tissue engineering, improving strategies for repair and regeneration.
References:
- Betancur, P., Bronner-Fraser, M. & Sauka-Spengler, T. Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proceedings of the National Academy of Sciences of the United States of America 107, 3570-3575, (2010).
- Le Douarin, N. The neural crest. (Cambridge University Press, 1982).
- Martinsen, B. J. & Bronner-Fraser, M. Neural crest specification regulated by the helix-loop-helix repressor Id2. Science 281, 988-991, (1998).
- Platt, J. B. Ectodermic origin of the cartilages of the head. Anat. Anz., 506-509, 1883.
- Simoes-Costa, M. S., McKeown, S. J., Tan-Cabugao, J., Sauka-Spengler, T. & Bronner, M. E. Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is Encrypted in the genome. PLoS genetics 8, e1003142, (2012).
- Simoes-Costa, M., Bronner, M.E. Reprogramming of avian neural crest axial identity and cell fate. Science 352, 1570-1573, (2016).
- Theveneau, E., Duband, J. L. & Altabef, M. Ets-1 confers cranial features on neural crest delamination. PloS one 2, e1142, (2007).


 (44 votes)
(44 votes)