The bumpy journey to the signal that kicks off endosperm development
Posted by Sarsim, on 9 September 2024
Here, Sara Simonini and Ueli Grossniklaus from the Institute of Plant and Microbial Biology and Zurich-Basel Plant Science Center, University of Zurich, tell the story behind the paper “A paternal signal induces endosperm proliferation upon fertilization in Arabidopsis”.
Fertilization is one of the most fascinating events during the development of an organism. In sexually reproducing multicellular organisms like animals and plants, fertilization involves the fusion of two gametes – a female egg cell and a male sperm. Gametes are highly specialized cells that, upon reaching maturity, await fertilization in a quiescent state. One way to achieve this is by inhibition of cell cycle progression, thus allowing gametes to arrest at a precise, stable stage. This aspect is crucial because uncontrolled gamete proliferation could have dramatic consequences, such as abortion of the progeny or a waste of resources.
Fertilization in plants is unique
When the egg cell and sperm fuse, their quiescent state is lifted and the cell cycle reactivated, so that the product of fertilization, the zygote, can initiate cell division. The molecular mechanisms that control the establishment of the quiescent state and its exit are still poorly understood.
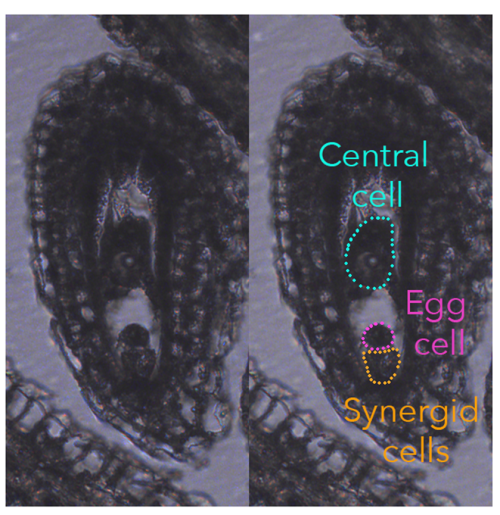
In flowering plants, the fertilization event is rather unique as they produce two types of female gametes, called egg cell and central cell. During the process of double fertilization, the two female gametes are fertilized by one sperm cell each, giving rise to the embryo and endosperm, respectively, the latter being a placental-like, nourishing tissue that sustains embryonic growth.
Typically, the egg cell and central cell derive from consecutive mitotic events of the same haploid megaspore, making them genetically identical. However, despite their genetic similarity, the egg cell and central cell have distinct identities, unique transcriptomes, different DNA contents (the central cell is homodiploid at maturity), and behave very differently once fertilized.
The fertilized egg undergoes morphological changes soon after fertilization. It progressively elongates, and its nucleus strongly polarizes towards the apical domain of the cell. The first cell division occurs approximately 20-24 hours after fertilization, resulting in an apical cell (forming the embryo proper) and a basal cell (forming the suspensor). In contrast, the fertilized central cell takes a different rhythm, committing its first division to initiate endosperm production already about 6-8 hours after fertilization.
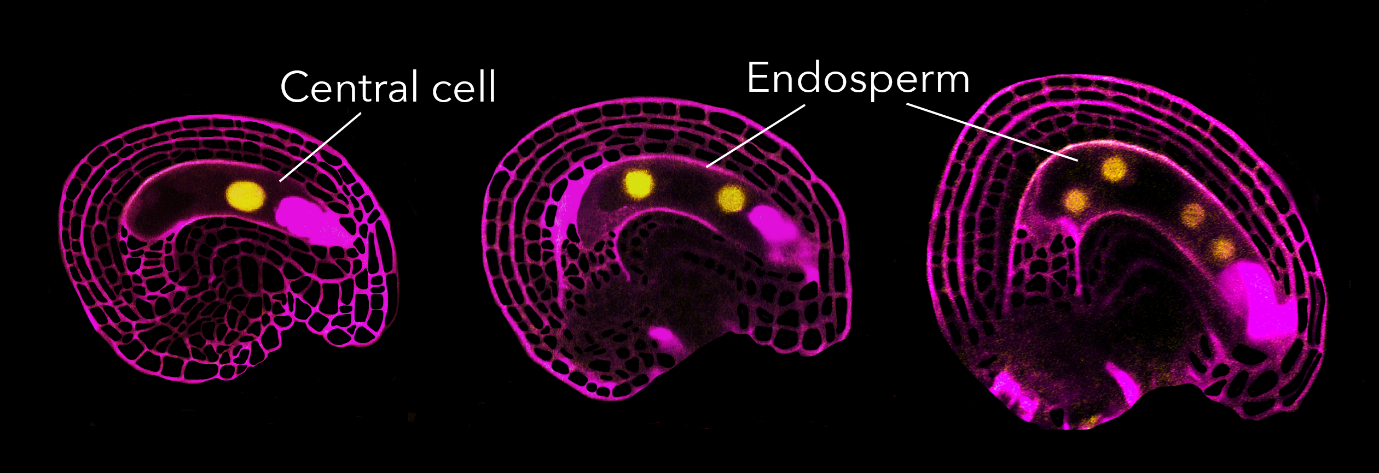
Cell cycle stage at which Arabidopsis gametes arrest
Over two decades ago, a hypothesis emerged suggesting the presence of a mechanism in the central cell that regulates the cell cycle, distinct from the one operating in the egg cell. The proposed idea was that a molecular brake prevents central cell division, and that fertilization acts as a trigger to release this brake, allowing division. This hypothesis stemmed from the observation of the rapid proliferation of the central cell after fertilization, as well as from the phenotypes exhibited by certain mutants where the central cell either divides in the absence of fertilization or is unable to divide once fertilized.
To understand fertilization’s impact on central cell quiescence, we initially determined the cell cycle stage at which the mature female gametes arrest. Quantifying DNA content in the female gametes is quite challenging as they cannot be collected in sufficient quantity for conventional ploidy analysis, such as flow cytometry. Our approaches involved propidium iodide staining to quantify DNA content, for which a reliable protocol was already established, and the imaging of histones tagged with fluorescent protein to infer the chromatin content in different nuclei of the ovule. These two approaches worked well and were reasonably straightforward. However, when it came to assessing DNA synthesis through nucleotide-analogue incorporation (EdU), well, we hit our head against a wall for about six months. It took a multitude of adjustments, trials, and a certain level of DIY attitude before we were able to establish a reliable, efficient protocol. But we made it!
It took a multitude of adjustments, trials, and a certain level of DIY attitude before we were able to establish a reliable, efficient protocol. But we made it!
The results of our ploidy analysis were both surprising and exciting. While we could confirm that the egg cell arrests in G2 as previously suggested, the central cell presented a completely different story. Its ploidy and behaviour suggested that its DNA synthesis (S phase) had initiated but not finished, and we could observe that fertilization was necessary for the S phase to be completed.
Finding the brake
Now that we knew the central cell is arrested in S phase, we wanted to identify the factor causing this arrest in DNA synthesis. Almost immediately, we considered RBR1, because it is a conserved cell cycle inhibitor known for regulating entry and progression through S-phase, and its absence causes central cells to proliferate in the absence of fertilization. The first confirmation that indeed RBR1 was our candidate came during a day at the microscope, observing the dynamics of an RBR1-YFP fusion protein during fertilization. For this type of experiments, we emasculated almost ready-to-bloom flowers by removing the stamens, so that self-pollination was avoided. The next day, we pollinated the pistils, marking the “0” time point. Then, after 4, 6, 8, 10, or 12 hours after pollination, we dissected the pistils and imaged the ovules using a multiphoton microscope. Normally, we pollinated between 8 and 9 in the morning, meaning that we had to spend quite some evenings at the microscope.
During these observations, we noticed that some central cells showed a RBR1-YFP signal, while others did not. After confirming the homozygosity of the RBR1-YFP line, it became evident that RBR1-YFP disappeared from the central cell only in fertilized ovules. This led us to the conclusion that something was degrading RBR1 at fertilization. Therefore, RBR1 acted as the brake, and fertilization somehow triggered RBR1 degradation, allowing the cell cycle to proceed.
Searching the signal that releases the brake
Just shortly after observing the turnover of RBR1 during fertilization, we received the sequencing results of transcriptomes from central cells at different time points before and shortly after fertilization that we had isolated by Laser-Assisted Microdissection (LAM). In practical terms, this technique allows us to isolate single cells from fixed, paraffin-embedded, and sliced tissues of interest. Completing this experiment took almost a year and a half for various reasons. The first significant obstacle was the global pandemic. We had just started to collect material when the institute went into a complete lockdown for about eight weeks, which meant that we lost at least two plant generations. The re-start was problematic too, because we had to do shifts to prevent overcrowding the labs, and experiments proceeded rather slowly. The second challenge was the time required make semi-thin sections of the material used for LAM. It takes approximately five days to gather enough material for a single replicate; our analysis covered four developmental time points, each performed in triplicate.
However, the results justified the long waiting time. Among the cell cycle-related genes potentially involved in RBR1 degradation, one D-type cyclin, CYCD7;1, caught our attention. Its expression peaked just around the moment when RBR1 is degraded in the central cell. Moreover, the literature indicated that CYCD7;1 is expressed only in stomata and pollen, and its ectopic expression in the female gametophyte was previously shown to induce proliferation of the unfertilized central cell. This led us to hypothesize that CYCD7;1 is paternally produced and stored in the sperm cells, and only upon fertilization, would CYCD7;1 be present in the same place and at the same time as RBR1, triggering its degradation. Observing CYCD7;1 messenger RNA location and delivery, as well as CYCD7;1 protein dynamics, confirmed our hypothesis. We also found that ectopic expression of CYCD7;1 in the central cell was sufficient to trigger RBR1 degradation and central cell division.
The only missing element was a visible phenotype. Mutant lines for CYCD7;1 (T-DNA and CRISPR-Cas9) were growing, and I (Sara) was confident in predicting the cycd7;1 mutant phenotype: paternal-effect seed abortion. This means that seeds would fail to develop when cycd7;1 mutant pollen was used as a male in a cross with a wild-type plant. Because RBR1 wouldn’t be degraded, the central cell wouldn’t divide, and no endosperm could be produced. However, upon inspecting the first cycd7;1 siliques under the microscope to evaluate the level of seed abortion, the result was hard to accept. All four cycd7;1 mutants I analysed exhibited a perfectly fine seed set – no seed abortion. We accepted the disappointing result that absence of paternal CYCD7;1 did not impact seed development. We went back to the LAM transcriptome, searching for alternative candidates, and stopped working on CYCD7;1. Sometime later, Ueli and I were having a meeting to discuss new hypotheses and strategies to further develop the project. As we revisited the CYCD7;1-related data, Ueli asked me which seed developmental stages I had been looking at for the phenotypical analysis, and he added “Do it again, look closer to the moment of fertilization”.
As we revisited the CYCD7;1-related data, Ueli asked me which seed developmental stages I had been looking at for the phenotypical analysis, and he added “Do it again, look closer to the moment of fertilization”.
That very afternoon, I sowed all the plant lines, and six weeks later, I made reciprocal crosses between wild-type and cycd7;1 plants again. This time, instead of looking at fully grown siliques, I sampled seeds at 12 hours after pollination, and the phenotype was evident: seeds generated by cycd7;1 pollen had fewer – or even no – endosperm nuclei compared to those derived from wild-type pollen. This meant that paternal delivery of CYCD7;1 is required for normal central cell division after fertilization. Central cells that receive a sperm cell lacking CYCD7;1 are blind to the fertilization event and do not divide immediately as they should. However, cycd7;1 mutant had no seed abortion, meaning that seed development can proceed normally even in absence of CYCD7;1 and, indeed, at 24 hours after pollination, cycd7;1-derived seeds showed endosperm proliferation. How can this happen? We hypothesized that other D-type cyclins, expressed from the maternal and/or paternal genome soon after fertilization, might compensate for CYCD7;1’s absence. This hypothesis turned out to be correct as we were able to delay endosperm proliferation even further when using pollen from plants mutated for four D-type cyclins.
Our results have not only addressed the fundamental question of how a cell determines the appropriate timing for division, but have also uncovered new and intriguing research directions. These include the understanding of how the central cell can arrest in S-phase, elucidating the mechanisms by which the CYCD7;1 messenger RNA is selectively stored in the sperm nucleus without degradation, and exploring the broader question of which other paternal or maternal signals regulate cell cycle arrest and progression in gametes. It also taught us the important lesson of formulating the right biological questions and designing the right strategies to address them. This is especially important when looking at developmental transitions, growth progression, and developmental processes in general: we cannot look at development if we do not take into consideration the time factor.


 (3 votes)
(3 votes)