The future of model organism research
Posted by Laura Hankins, on 1 December 2022
For permission to reproduce, contact the MRC National Mouse
Genetics Network.
The September issue of our journal Disease Models & Mechanisms (DMM) has a focus on how researchers can best leverage model organisms to improve our understanding of human health and disease. Here, we highlight some of the articles and encourage you to visit the journal website, where all the articles are available to read Open Access.
The issue includes a Special Article by Keith Cheng, Hugo Bellen and colleagues on the importance of promoting validation and cross-phylogenetic integration in model organism research. The article draws on a discussion series organised by the National Institutes of Health (NIH) to gather opinions on how researchers can further extend the utility of model organisms in the future. The authors discuss some of the key ideas identified by the NIH discussions, including:
- Developing new tools and technologies.
Ideas include humanising model organisms to better model human diseases, generating reference atlases to allow comparison between model organism and human data, and developing new reagents such as species-specific antibodies. - Broadening the range of model organism species used in research.
Different species offer specific advantages for modelling human disease. For example, hamsters share similar lung physiology to humans so have proved useful in COVID-19 studies. Expanding the range of species used in disease research could therefore offer researchers greater access to useful models that have been previously overlooked or underused. - Increasing genetic variation.
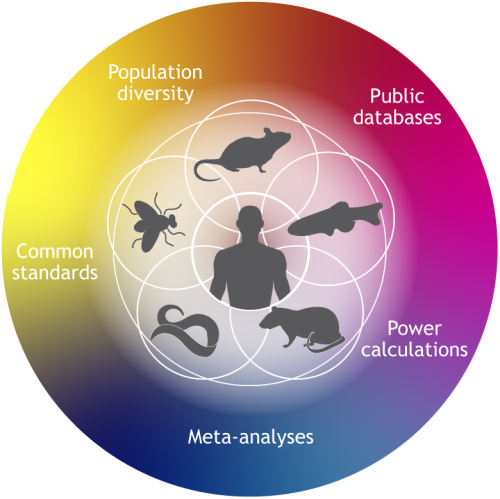
Laboratory animal strains are often inbred to reduce variation. However, humans have a great deal of genetic diversity, so it will be important to use a suite of diverse models to better reflect the variation in human disease. - Integration between different disciplines and organisms.
It will be important to integrate phenotypic data from different disciplines (such as biochemistry, cell biology, genomics and behavioural studies) as well as data from studies in different model organism species. This will help to form a more complete picture of the disease phenotype.

An example of a new initiative that aims to improve integration across disciplines is the UK Medical Research Council (MRC) National Mouse Genetics Network. You can read more about this in Owen Sansom’s Editorial. Owen explains that the Network will comprise seven research clusters, each focusing on different yet complementary research areas. The aim is to share data, techniques and resources, and to encourage collaboration within the mouse model community.
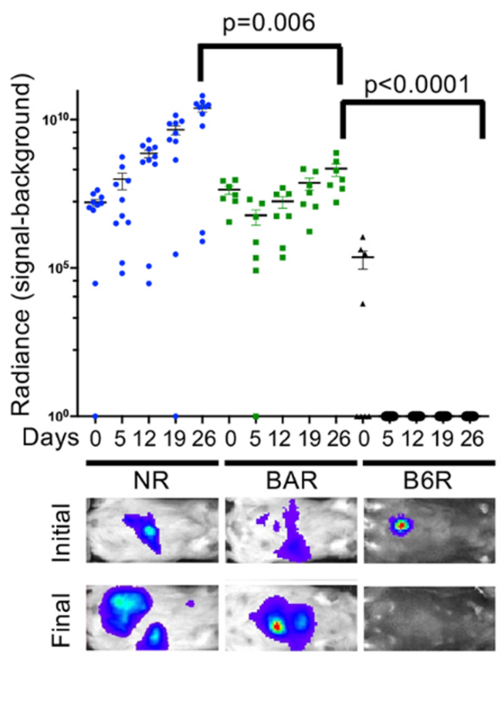
The September issue of DMM also contains a study from Muneer Hasham and colleagues at the Jackson Laboratory (JAX), USA, which expands the genetic diversity of mouse models available for xenografting experiments. Xenografting human cancer cells in mice is a well-established method for studying tumour development and testing potential therapeutic drugs. These mice must lack B- and T-lymphocytes for xenografting to be successful, and researchers can achieve this by generating mouse strains that are Rag1 deficient. However, the genetic diversity of available Rag1-/- mouse strains is limited.
In their paper, the JAX group generate five genetically diverse Rag1-/- mouse strains. Together, these strains cover 90% of the known allelic diversity in the mouse genome. They show that the genetic background of the host strain plays an important role in the outcome of the xenograft; for example, they find that tumour size varies between the five different host strains. This article was also highlighted as the DMM Editor’s Choice.
You can read a response to the work by Dr Hasham and colleagues in the same issue of DMM, where Ryan Devlin and Ed Roberts tackle the question of how researchers can build a healthy mouse model system to better interrogate cancer biology. Taking the JAX lab paper as a case study, they suggest implementing a ‘Swiss cheese model’ to design better experiments. This model is already used in accident prevention strategies, where the concept of layering multiple strategies (each with their own weaknesses, as represented by holes in the slice of cheese) is employed to reduce the chance of an accident.
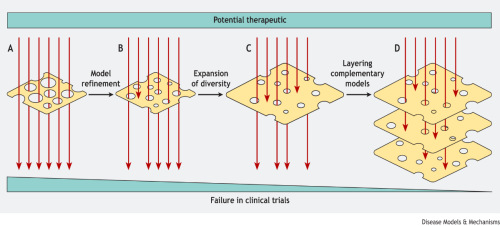
In a similar vein, Ryan and Ed suggest that researchers can layer a suite of different mouse models, rather than trying to identify the ‘best’ model. This is because each model has its own weaknesses so, by combining models that complement one another, scientists can reduce the chances of a harmful or ineffective chemotherapeutic drug reaching the clinical trial stage.
Finally, researchers from JAX have also provided a Perspective for the October issue of DMM. In the article, Karen Svenson and colleagues respond to the NIH’s 2021 recommendations to improve the reproducibility of animal studies. They share their own experiences of addressing this issue, with a specific focus on mouse models.
Want to keep up with the latest news from DMM? Visit the DMM website and follow the journal on Twitter and/or Mastodon to find out more.


 (No Ratings Yet)
(No Ratings Yet)