Traditional Embryology with Modern Imaging Approaches: Investigating Morphogenetic Feedback on Pattern Formation
Posted by Timothy Fulton, on 9 October 2020
Timothy Fulton, Vikas Trivedi, Andrea Attardi & Benjamin Steventon
As developmental biologists, we often find ourselves carefully looking at developing embryos as they undertake a dramatic and fascinating task: making a complex organism out of a few initial embryonic cells. On the other hand, for engineers, building complex structures out of simple constituent parts under a defined set of rules is what creates excitement every day. Together, the bioengineer and the embryologist wonder how cells behave in a way that shapes the animal, whilst simultaneously deciding on their fate. Clearly, these two processes of morphogenesis and pattern formation must occur in concert.
We know that vertebrate embryos achieve this by integrating cues and signals from the extraembryonic environment. Such cues do not act alone though, as they are coupled to an intrinsic ability of early cells to drive morphogenesis and patterning. This latter feature of embryonic cells, known as self-organisation, is remarkably highlighted when cells are separated from their extra-embryonic constraint. An example of this is given by mouse and human embryonic stem cells (mESCs/iPSCs). Under controlled culturing conditions, these cells aggregate as ‘gastruloids’ (van den Brink et al., 2014; Moris et al., 2020), groups of cells that perform and recapitulate essential aspects of axis specification and pattern formation seen in intact mouse or human embryos.
Such ideas about self-organization of embryonic cells were in the atmosphere during lab meetings and tea time discussions at the Department of Genetics. Vikas, who had recently joined the lab of Alfonso Martinez Arias as a postdoctoral fellow, came with the experience of having worked with zebrafish before and was wondering about a fish equivalent of gastruloids. At the same time, Andrea, a masters student in Ben Steventon’s lab, while working on a project to lineage trace Neuromesodermal Progenitors during gastrulation, was having similar thoughts (Attardi et al., 2018). The question arose quite spontaneously: could zebrafish embryonic cells tell us something new about morphogenesis and self-organisation, if we cultured them free of their embryonic constraints? That looked like a good opportunity for exploring what the collaboration of bioengineering and embryology could lead us to. We therefore decided to attempt culturing zebrafish embryonic cells at early stages of development, starting by explanting cells from the 256 cell stage embryo, prior to the midblastula transition. Strikingly, we observed that in doing so, these masses of cells elongated over a period of seven to ten hours. We observed this phenomenon in a range of media, with and without serum, including inert Ringer’s Solution. These results nicely echo those of Jane Oppenheimer (1936), who found similar results when explanting blastoderm from another teleost, Fundulus heteeroclitus.
Pescoid Elongating – Wildtype pescoid elongating in normal L15 + 3% FBS Serum imaged on a widefield microscope
After identifying optimal incubation conditions, Tim set about understanding which cell types formed within these elongating explants. After making many hundreds of pescoids, and using multiplex in situ Hybridisation Chain Reaction (Choi et al., 2018), he demonstrated that these explants produce all three germ layers in their elongated form. We observed the mesodermal marker tbxta expressed in the elongating pole, with sox17 marking the endoderm in the core of the explants. In the non-elongating end, we observed sox2 expressed marking the ectoderm. In addition to these marker genes, we also observed expression of germ cell markers nanos and vasa, as well as expression of the hindbrain marker gene krox20 which, as in the embryo, was expressed in two clearly defined stripes of expression in the non-elongating end. Interestingly, we did not observe expression of otx2 positive forebrain cell.
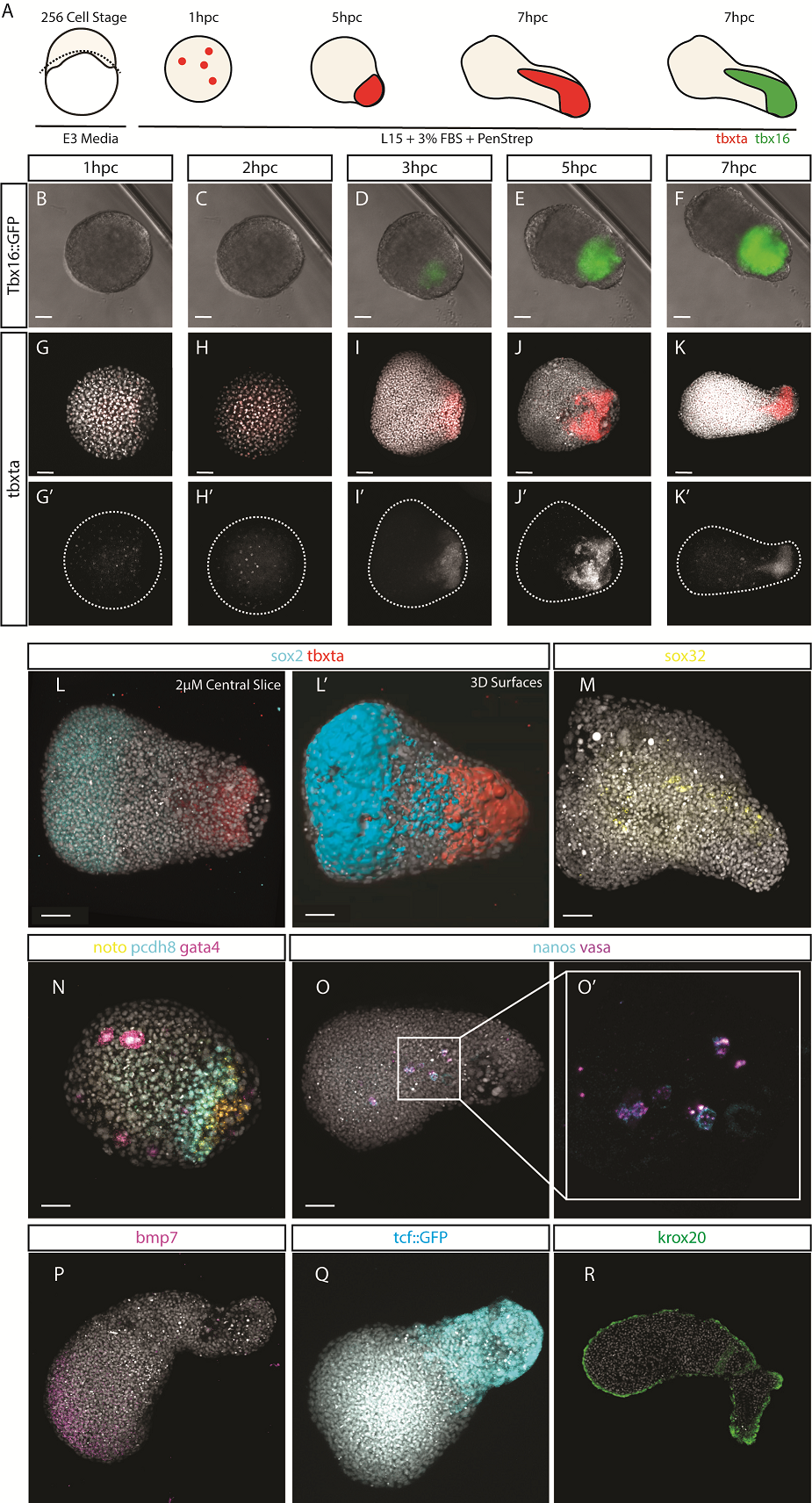
We wanted to test exactly how robust the germ layer formation was to mixing of the cells. In order to test this, we cut and totally dissociated and reaggregated the explants immediately after explanting using two eyelashes glued to capillary tubes. Tim and Ben swept the cells together gently under a microscope in order to break down and reassemble the explant. A very delicate procedure, made harder by Ben’s insistence that we listened to Test Match Special, in order to keep up to date with the Cricket World Cup! When we reaggregated pescoids, we observed that the explants rarely elongated, however the majority still expressed tbxta. We hypothesised that the initial size of the explant is important for elongation potential, as quartered explants rarely elongate, however half sized explants retain this potential.
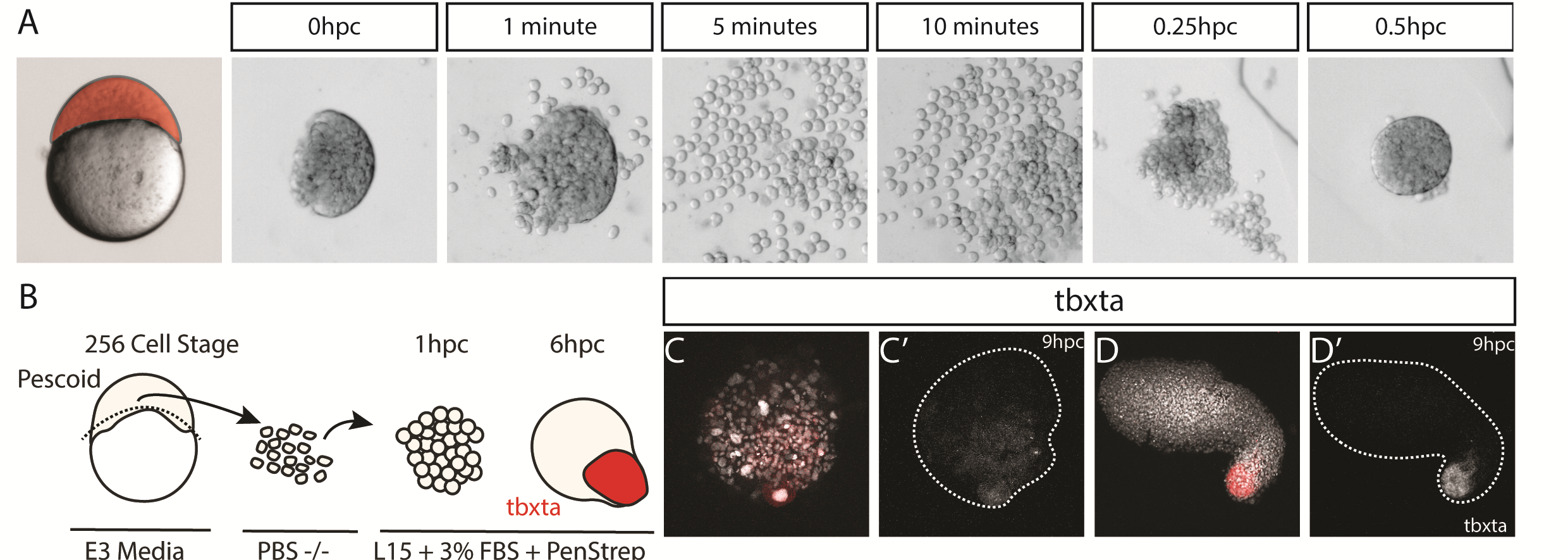
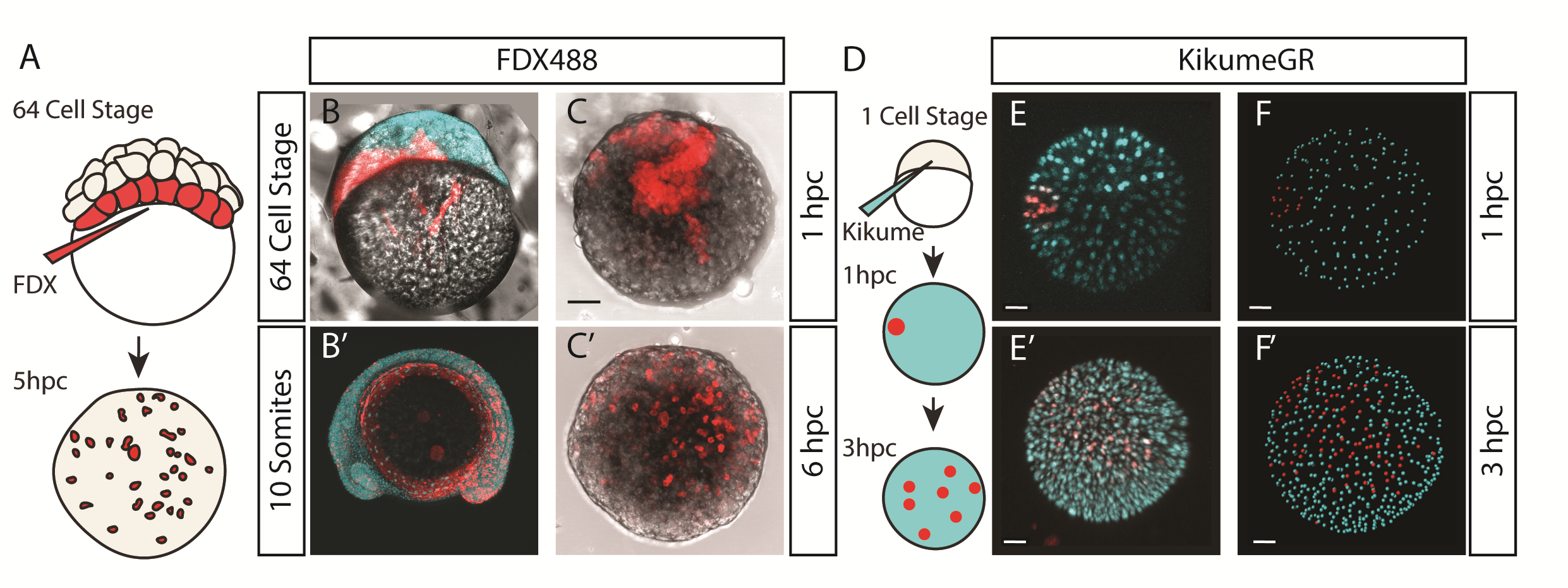
We were also interested to discover how much cell mixing was occurring in these explants naturally. Using two different methods, Chaitanya labelled either the edge of the explant, or the marginal cells of the embryo prior to explanting, and then tracked these cells post explant. In all situations, we observed that the labelled cells mix significantly with the non-labelled cells prior to elongation. Schauer et al. (2020) recently conducted very similar experiments to us, and demonstrated that the prepatterns provided to the embryonic cells by the yolk prior to the stage at which we take explants are essential for the formation of the elongation. We also found that the whole process is dependent on early Nodal activity, a result that fitted nicely with the work of (Williams and Solnica-Krezel, 2020).
Nodal in Explants – Activin Response Element::GFP reporter for Nodal activity demonstrating correlation of Nodal signalling with the elongation
We had some excellent discussions with Alexandra Schauer at the EDBC meeting in Alicante in 2019 and really enjoyed being able to share and discuss our findings with one another. However, the extensive mixing we observe here demonstrates that whatever the mechanism between prepattern and pattern formation is, it must also be highly robust to cell mixing over long timescales of 3 to 5 hours. Overall the individual cells regularly swap positions with one another, however, at the organism level, the explants form reproducible domains of stable gene expression.
How these different domains of gene expression came about remained unclear, and to investigate this, we looked at the signalling events taking place within the explant. We observed that initially the activity of Wnt and BMP signalling completely overlapped. During elongation however, these two activity domains separate, with Wnt activity being restricted to the elongation, and BMP to the non-elongating end. We hypothesized that the elongation may be the driving force behind the separation of these two domains, and by inhibiting morphogenesis both genetically and pharmacologically, we demonstrated that this is indeed the case; without morphogenesis, the two domains remain overlapping and two krox20 stripes were never observed. The explants however are unable to produce an environment with very low levels of both Wnt and BMP, and therefore this may explain why Oxt2 positive cells are not produced.
Pescoids Elongating with TCF::GFP and BRE::RFP. TCF::GFP reporter for Wnt activity, demonstrating Wnt signalling occurs across the entire explant their localises to just the elongation (Green to White). BMP Response Element fusion of RFP demonstrates BMP signalling is active across the entire explant then localises to just the non-elongating end.
We therefore had a situation where our explants are displaying a very high level of cell mixing, with cells regularly changing positions with one another, whilst also maintaining distinct regions of gene expression. Whilst our work here begins to answer how these domains form, it does not answer how these domains are then maintained. This opens an intriguing question: how do cells, via signalling and gene regulatory networks, update the cell’s positional information whilst constantly moving?
Traditionally, this type of question would invoke ideas of the French Flag Model, which describes a situation where morphogen gradients inform cells of their position and, therefore, of their fate decision. However, as Wolpert described in the French Flag Problem (Sharpe, 2019; Wolpert, 1969), this idea becomes difficult to apply when cells also regularly change positions with one another and therefore scramble the pattern as it is being produced. This is something which Tim is continuing to investigate as part of his PhD. Using live imaging data and tracking of individual cells, we are overlaying dynamical systems models of a GRN onto in vivo tracks in order to model cell fate decisions in a tissue undergoing active morphogenesis. The Steventon Lab is also interested in investigating how pattern formation may be an emergent property of gene regulatory networks and morphogenesis working together. Using the idea of tectonic plates, the term tissue tectonics (Busby and Steventon, 2020) describes how morphogenetic movements regulate the timings of interactions between different tissues as they slide together or apart, similar to as we see in these pescoid explants, and other embryonic systems. The Trivedi lab is pursuing studies on understanding the biophysics of self-organization that leads to patterning in early embryonic systems including pescoids.
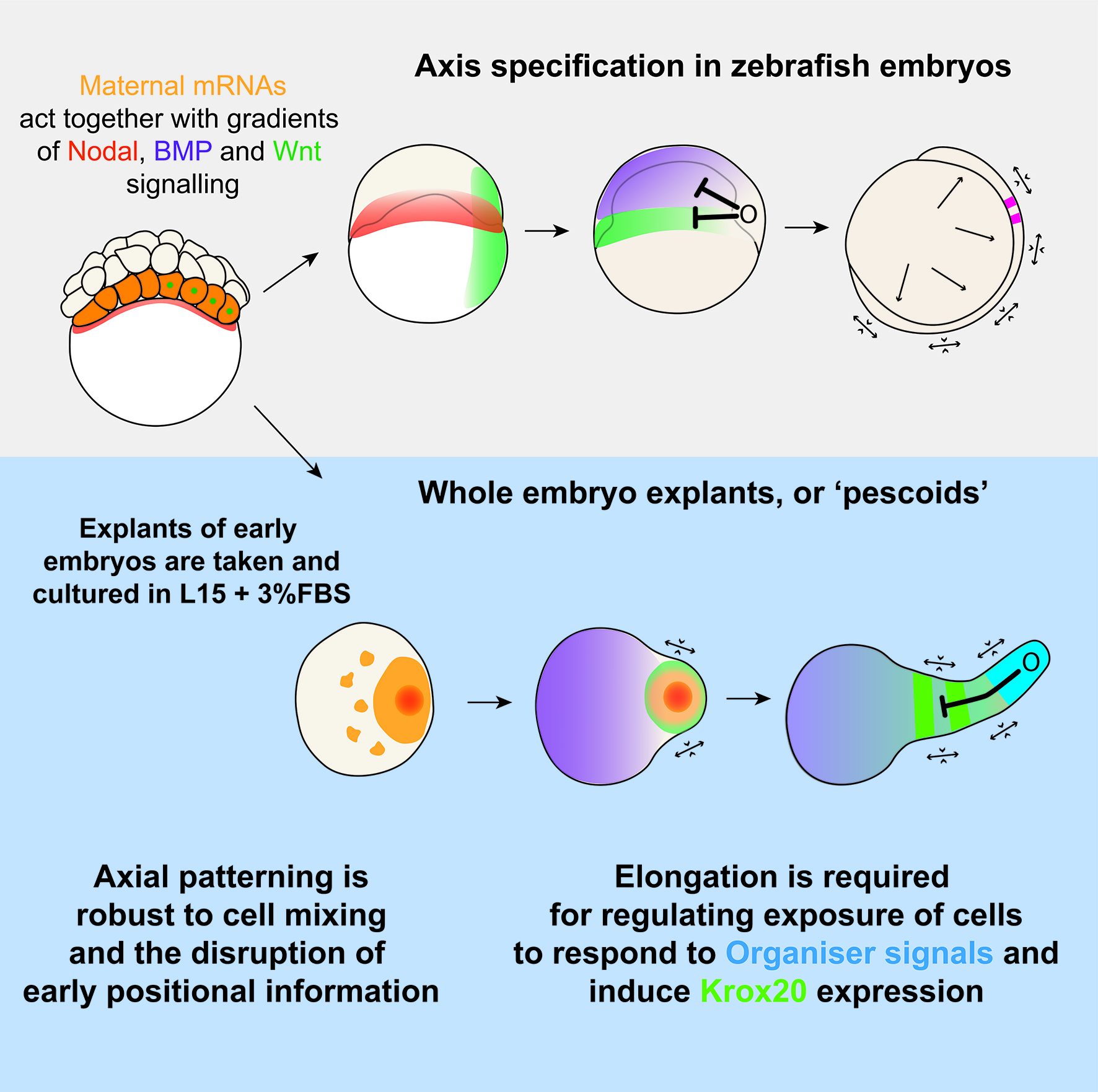
Overall, this work, and the ideas of tissue tectonics propose a model where pattern formation is as much the output of genes and local cell rearrangements, as it is the more global tissue movements which shift cells into and out of range of one another. By considering patterns as an emergent property of morphogenesis and GRNs, we can begin to ask questions about how these components are brought together, and what role cell-cell signalling plays in this.
Read the full paper in Current Biology
References
Sharpe, J. (2019). Wolpert’s French Flag: what’s the problem? Development 146.


 (2 votes)
(2 votes)