What might evolutionary muscle loss and pathological atrophies have in common?
Posted by maip.tran, on 8 January 2020
By Mai P. Tran and Kimberly L. Cooper
“It’s the cutest rodent I have ever seen, even cuter than a cuddly hamster, and it would be fun doing a rotation for the opportunity to work with this animal.” That was my thought, as a first-year graduate student, when I first heard Kim present her research on the development of digit loss in the jerboa. I was completely hooked by the jerboa’s fascinating biology, and the Cooper lab became my second family. I had no idea then how working with the challenges of a non-traditional species would also mature my scientific thinking.
The vertebrate form is incredibly diverse, as evident not only in the body shapes and sizes but also in the limbs. For example, bats have elaborated forelimbs capable of flight while snakes have lost all four limbs. Such diversity has long inspired scientists and provided opportunities to investigate both developmental and evolutionary processes. To understand the developmental programs that explain limb diversity, the Cooper lab uses the bipedal jerboa (Figure 1), a close relative of the mouse with very different hindlimbs. The jerboa hindlimb has lost the first and fifth toes, fused the three central metatarsals into a single bone, disproportionately elongated the foot, and lost all intrinsic foot muscles. These modifications of the limb form allow the jerboa to jump high and rapidly change orientation, an advantage to escape predators in its desert environment with sparse vegetation. The jerboa and mouse have similar forelimb morphology, which serves as a great internal control for comparative anatomy and genomics to identify potential mechanisms specific to hindlimb divergence.

Our recent work, Tran et al. eLife 2019, showed that the normal developmental process of muscle loss in the jerboa is similar to pathological muscle atrophy. In this post, I will elaborate on our findings, sprinkled with scientific speculation, and discuss the experimental challenges that we encountered during our exploration of the fascinating biology of this non-traditional laboratory animal model.
In 1979, Susan Berman, then a graduate student at the University of Pittsburgh who later became faculty at College of the Holy Cross, published a description of the hindlimb skeletal and muscular anatomy of many bipedal rodents including several species of jerboas1. When I started my work in 2014, we knew from her work that the adult jerboa has no ‘intrinsic’ muscles within its feet that spread and draw together toes of more basally branching species. Instead, most jerboa species have robust tendons in each location that we presume stabilize the ankle when animals jump and land with great force. However, we knew nothing of the developmental process leading to the evolutionary loss of these muscles.
Jimmy Hu, a graduate student with Cliff Tabin while Kim was a postdoc, had recently shown that limb myoblasts require reception of Shh signal to complete migration into the distal limb, as Smoothened receptor loss-of-function in mouse muscle causes a complete absence of muscles in the hands and feet2. A similar failure of muscle progenitor cells to populate the distal jerboa hindlimb seemed a simple explanation for the adult phenotype. We thought this project might be ‘low-hanging fruit’ for our nascent lab. We were therefore surprised to find that not only do myoblasts reach the foot, they fuse, differentiate, and form sarcomeres before rapidly disappearing in the first week after birth.
We next assumed the muscles must have died. Programmed cell death eliminates ‘extra’ tissue to shape developing structures in many contexts, including the process that carves interdigital webbing away from the fingers and toes like pulling dough away from the edges of a cookie cutter. Convinced this must also be the case for developing muscle that disappears so quickly, I performed multiple assays at all stages of muscle loss fruitlessly searching for signs of compromised cell membrane, fragmented DNA, or immune cell recruitment. I’ve also looked at thousands of muscle cells by transmission electron microscopy, a classic approach used to describe programmed cell death before there were kits and antibodies. Although myofibers don’t look entirely normal (they have large lipid droplets and vacuoles, and the sarcomere disintegrates), the plasma membrane appears intact, the nucleus appears normal, and there is no immune cell infiltration suggesting these cells do not die. I thought that I must be doing something wrong, but I had included all the positive and negative controls I could think of and covered all the stages of muscle loss. The more data I amassed suggesting this simple hypothesis was invalid, the more frustrated I grew. In contrast, my advisor became more and more excited.
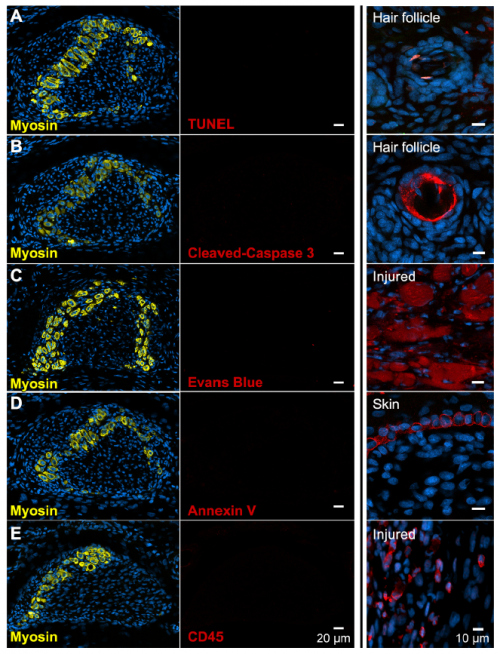
If muscle cells did not seem to die, what was their fate? We considered the possibility that muscle cells switch identity, which would explain the disappearance of muscle proteins. The robust tendon in the adult and the appearance of collagen-producing cells surrounding odd extra-cellular aggregates of muscle filament protein led us to hypothesize that perhaps muscle cells could become some type of collagen-producing connective tissue fibroblast.
To definitively show that muscle cells become another cell type, or that they die by an unusual mechanism we were unable to detect by common assays, we would need to lineage label jerboa foot muscle cells and follow their existence over time. For over three years, we tried viral infection and plasmid electroporation approaches using multiple ‘muscle-specific’ promoters to lineage trace muscle cells using a Cre/lox reporter system, but none was successful. There were two technical complications hindering our ability to successfully lineage label muscle cells, discussed briefly in the response to eLife reviewers; we were unable to label a sufficient number of cells in neonatal mouse or jerboa feet, and none of the promoters specifically labeled muscle in either species. We hope that our ongoing work utilizing single-nucleus sequencing of muscle and surrounding tissue can help shed light on the fate of muscle cells by reconstructing a ‘virtual lineage’ using global similarities in the transcriptome of heterogeneous cell types3. Though not as definitive as a lineage tracing experiment, this might provide valuable insight into the most likely fate of these cells.
During those years of trying to determine the fate of the muscle, I also took a parallel approach to learn more about the developmental trajectory of jerboa foot muscles and to identify the earliest signs of impending loss. I found that muscle sarcomeres, though assembled, never fully mature to align with one another and instead became disorganized into pools of filaments. We were curious to know if the process of muscle disassembly happened all at once or in some orderly fashion. My observation that Desmin was the first protein to lose its striated pattern was our first break in this challenging project, since this was similar to what was observed in conditions of skeletal muscle atrophy. Even though atrophy is typically considered a response to disease or injury, might it be possible that the ‘normal’ developmental process of jerboa foot muscle loss is similar to atrophy? Indeed, expression of genes associated with pathological muscle atrophy, called “atrogenes,” was significantly higher in jerboa foot muscles than other ‘typical-developing’ muscles.
In 1883, Daniel John Cunningham, a Scottish physician, zoologist, and anatomist, reported the presence of intrinsic foot muscles in fetal horses, deer, and oxen that lack these muscles as adults. Cunningham observed fat and fibrotic tissue infiltration that replaced the disappearing foot muscles4. When we re-read the work of Cunningham as we prepared our own manuscript, we realized that Cunningham had left a hint we weren’t initially prepared to appreciate when he wrote “it is an instance of pathological change assisting a morphological process”4. Now aided with molecular and genetic tools, we show that the natural process of muscle loss in jerboa does indeed share molecular characteristics with pathological atrophy. If we look carefully at other ‘non-traditional species’, we may find that foot muscle atrophy in the jerboa is just one of many cellular responses associated with injury or disease in humans that is utilized in the normal development and physiology of other species.
While it is tempting to think that we might prove a cause and effect relationship by knocking down expression of the atrogenes to rescue jerboa foot muscle, it is important to consider differences between macroevolutionary phenotypes and mechanistically simpler phenotypes that are more typically studied in the lab. A comparison of the foot anatomy of living jerboas and their nearest relatives reveals that a subset of muscles was lost about 20 million years ago gradually leading to absence of all foot muscle in more recently derived jerboas. We therefore expect that multiple genetic changes contribute to the mechanism of muscle cell loss, perhaps including mutations that primarily affect integrated tissues like the skeleton and connective tissues. Although multiple genes responsible for evolutionary phenotypes can be identified by quantitative trait locus (QTL) analyses, the approach requires that species with different morphologies produce fertile hybrids. The jerboa lineage diversified too long ago for such mattings to be possible. Instead, we are currently applying interspecies differential RNA-Seq of mouse and jerboa foot and forearm muscles to identify genes with expression differences that are specifically associated with jerboa foot muscle loss. In addition to building a comprehensive molecular profile of this process, these data provide an opportunity to identify genes not previously known to influence muscle maturation and maintenance.
Thinking about the big picture of my research brought me to the realization that evo devo isn’t simply the study of individual phenomena in unusual critters. All deviations from ‘typical development’, both adaptive and maladaptive, provide insight into unifying principles. Studies of non-traditional species, while they can be technically challenging, will not only reveal the mechanisms of evolutionary malleability, but also advance our understanding of fundamental biological processes.
References
- Berman, S. L. Convergent evolution in the hindlimb of bipedal rodents. Journal of Zoological Systematics and Evolutionary Research23, 59–77 (1985).
- Hu, J. K.-H., McGlinn, E., Harfe, B. D., Kardon, G. & Tabin, C. J. Autonomous and nonautonomous roles of Hedgehog signaling in regulating limb muscle formation. Genes & development26, 2088–102 (2012).
- Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature Biotechnology 32, 381–386 (2014).
- Cunningham, D. J. The Development of the Suspensory Ligament of the Fetlock in the Foetal Horse, Ox, Roe-Deer, and Sambre-Deer. Journal of anatomy and physiology18, i1-12 (1883).


 (2 votes)
(2 votes)