GATA6 and the power of single cells
Posted by Nestor Saiz, on 29 May 2014
Any mammal who celebrated Mother’s Day earlier this month realizes how important mothers are for us and the tight bond between them and their children. Forget clean shirts and packed lunch every day; for us developmental biologists, there is no better reflection of this bond than the extraembryonic membranes that support the growth of the fetus in the uterus. These tissues, among other roles, serve as circulatory, digestive and excretory systems until we develop our own. As a matter of fact, they are so important that they begin to form even before the fetus itself – during the preimplantation period, before the embryo attaches to the uterus (check out the cartoon below). Therefore, the very first decisions cells need to make during a mammal’s life are whether to become part of the extraembryonic lineages (called trophectoderm and primitive endoderm at this stage) or become the foundation of the fetus (the epiblast).
Historically, the first of these decisions (whether or not to become trophectoderm) has received more attention by researchers. However, the second one, whether to become primitive endoderm or the pluripotent, embryonic epiblast, has only been studied in more detail over the last decade or so. A number of molecular markers and the cellular behaviors involved in this process have been described, although no transcription factor has yet been shown to be essential for primitive endoderm specification. In the Hadjantonakis lab, we have recently looked at the role of the main suspect – the GATA family transcription factor GATA6 – and we are publishing the results in the current issue of Developmental Cell [1].
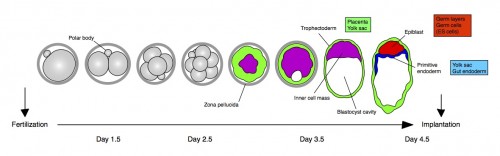
This study is important for two reasons: one biological and the other one technical. As I mentioned earlier, this is the first study to show a transcription factor that is absolutely required for primitive endoderm specification. Gata6 null embryos completely lack primitive endoderm because all cells of the inner cell mass default prematurely to epiblast fate (the future fetus). Although they show no other phenotypic defect, these embryos die upon implantation, presumably because there is no tissue to support the growth of the epiblast. We also show that Gata6 heterozygous null embryos have reduced levels of GATA6 and, as a consequence, show a delay in primitive endoderm specification. Finally, this study clarifies the love triangle formed by GATA6, the pluripotency champion NANOG and the fibroblast growth factor (FGF) signaling pathway. FGF4 (or basic FGF/FGF2) is the instructive signal for primitive endoderm specification among inner cells in the blastocyst; however, as we show for the first time in this paper, GATA6 is necessary for cells to respond to FGF4, consequently downregulate NANOG, and acquire primitive endoderm identity. Our paper not only provides a mechanism for the induction of primitive endoderm fate, but also suggests that the relative levels of GATA6 to NANOG in each cell could tip the balance towards either an extraembryonic or an embryonic fate. How these levels change and how cells can measure them remains to be seen…
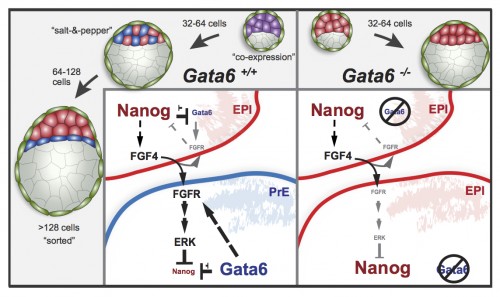
Our study is also exciting because of the approach we used to analyze the Gata6 allelic series. For those less familiar with it, the mouse preimplantation embryo is as good a model for (live) imaging and single-cell analysis as it is unfit for biochemistry. It is very small, with few cells, and can develop in vitro, without maternal support, in standard culture conditions. This means we can image and analyze the behavior of every single cell within the embryo using a rather simple experimental setup, making it an ideal model to study mammalian cell differentiation in situ. Despite these features, only recently have there been attempts at quantifying gene expression and doing single cell analyses, partly fueled by the technology that has become available [see references 2-4, or, more recently, 5-7, for examples].
In this work, we have used MINS, a piece of software recently developed in our lab [8] that can segment nuclei in series of confocal images with higher accuracy and lower manual input than any other software we have tried so far – including popular (and powerful) solutions like ImageJ or Imaris. MINS collects spatial coordinates and fluorescence intensity data for every channel on each cell, gives them a unique ID, and generates a data matrix for each embryo. After some manual curation for under/oversegmentation and to remove the occasional outlier (generally dead cells), the data is ready for you to do maths and statistics to your heart’s content. Being biologists, large data matrices send shivers down our spines, so we teamed up with physicist-turned-biologist Stefano Di Talia and, lo and behold, the numbers began to make sense! Furthermore, because MINS also collects positional information, we were able to relate changes in protein levels and cell identity to position within the embryo. To a certain extent, this kind of analysis allows us to bypass Western Blots and study protein expression in individual cells, while preserving positional information, which is lost when cells are disaggregated for gene expression analyses. This study is the first of several from our lab where we will apply this pipeline to three- and four- (time lapse) dimensional datasets to understand better the earliest cell fate decisions in mammalian development. We hope the implementation of this analysis pipeline by other labs will improve it and help generate even higher quality data in future studies. The combination of these advanced algorithms for image analysis with single-cell expression profiling techniques will let researchers study with high resolution the molecular mechanisms controlling cellular processes in situ, in intact tissues or embryos. For further proof that this is becoming a thing, have a look at these recent comments: 9, 10.
If you want to discuss or comment, do it below or you can reach me on twitter
References:
1.Schrode, N., Saiz, N., Di Talia, S., & Hadjantonakis, A. (2014). GATA6 Levels Modulate Primitive Endoderm Cell Fate Choice and Timing in the Mouse Blastocyst Developmental Cell, 29 (4), 454-467 DOI: 10.1016/j.devcel.2014.04.011
2.Kurimoto, K., Yabuta, Y., Ohinata, Y., Ono, Y., Uno, KD., Yamada, R., Ueda, H., & Saitou, M. (2006). An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis Nucleic Acids Research, 34 (5) DOI: 10.1093/nar/gkl050
3.Plusa, B., Piliszek, A., Frankenberg, S., Artus, J., & Hadjantonakis, A. (2008). Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst Development, 135 (18), 3081-3091 DOI: 10.1242/dev.021519
4.Guo, G., Huss, M., Tong, G., Wang, C., Li Sun, L., Clarke, N., & Robson, P. (2010). Resolution of Cell Fate Decisions Revealed by Single-Cell Gene Expression Analysis from Zygote to Blastocyst Developmental Cell, 18 (4), 675-685 DOI: 10.1016/j.devcel.2010.02.012
5.Frum, T., Halbisen, M., Wang, C., Amiri, H., Robson, P., & Ralston, A. (2013). Oct4 Cell-Autonomously Promotes Primitive Endoderm Development in the Mouse Blastocyst Developmental Cell, 25 (6), 610-622 DOI: 10.1016/j.devcel.2013.05.004
6.Le Bin, G., Munoz-Descalzo, S., Kurowski, A., Leitch, H., Lou, X., Mansfield, W., Etienne-Dumeau, C., Grabole, N., Mulas, C., Niwa, H., Hadjantonakis, A., & Nichols, J. (2014). Oct4 is required for lineage priming in the developing inner cell mass of the mouse blastocyst Development, 141 (5), 1001-1010 DOI: 10.1242/dev.096875
7.Ohnishi, Y., Huber, W., Tsumura, A., Kang, M., Xenopoulos, P., Kurimoto, K., Oleś, A., Araúzo-Bravo, M., Saitou, M., Hadjantonakis, A., & Hiiragi, T. (2013). Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages Nature Cell Biology, 16 (1), 27-37 DOI: 10.1038/ncb2881
8.Lou, X., Kang, M., Xenopoulos, P., Muñoz-Descalzo, S., & Hadjantonakis, A. (2014). A Rapid and Efficient 2D/3D Nuclear Segmentation Method for Analysis of Early Mouse Embryo and Stem Cell Image Data Stem Cell Reports, 2 (3), 382-397 DOI: 10.1016/j.stemcr.2014.01.010
9.Wen, L., & Tang, F. (2014). Reconstructing Complex Tissues from Single-Cell Analyses Cell, 157 (4), 771-773 DOI: 10.1016/j.cell.2014.04.024
10.Robson, P. (2014). Deciphering Developmental Processes from Single-Cell Transcriptomes Developmental Cell, 29 (3), 260-261 DOI: 10.1016/j.devcel.2014.04.032


 (5 votes)
(5 votes)