BSDB Gurdon Summer Studentship Report (12)
Posted by BSDB, on 24 October 2017
![]() Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding.
Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding.
Our first report from the 2017 group of student awardees comes from Josie Elliott (student at University of Oxford), who undertook her studentship with Alison Woollard at the Dept. of Biochemistry in Oxford.
What is the role of wrt-2 and wrt-4 in left-right asymmetry in C.elegans?
 Once upon a time a genetic screen identified a signalling pathway that caused Drosophila melanogaster embryos to develop a ‘lawn’ of denticles rather than forming them only at parasegment boundaries. Thus the so-called Hedgehog signaling pathway was born (Nusslein-Volhard & Wieschaus, 1980). This pathway has revealed itself to be one of the core signal transduction pathways in regulating animal development. Through gene duplication and diversification events, different types of hedgehog proteins have been found across vertebrates. The brilliantly named Sonic Hedgehog ligand is the best studied ligand present in mammals. Loss of expression of Sonic Hedgehog in mice causes defects in left right asymmetry (Levin et al., 1995).
Once upon a time a genetic screen identified a signalling pathway that caused Drosophila melanogaster embryos to develop a ‘lawn’ of denticles rather than forming them only at parasegment boundaries. Thus the so-called Hedgehog signaling pathway was born (Nusslein-Volhard & Wieschaus, 1980). This pathway has revealed itself to be one of the core signal transduction pathways in regulating animal development. Through gene duplication and diversification events, different types of hedgehog proteins have been found across vertebrates. The brilliantly named Sonic Hedgehog ligand is the best studied ligand present in mammals. Loss of expression of Sonic Hedgehog in mice causes defects in left right asymmetry (Levin et al., 1995).
The story was expanded into C.elegans by bioinformatics (Burglin, 1996). A search of the C.elegans genome for homologues of the carboxyl terminal ‘hog’ domain revealed a family of proteins which did not contain the amino terminal ‘Hedge’ domain but instead a novel domain dubbed ‘Wart’. Thus the warthog (wrt) genes were named. Further genome analysis revealed genes which contained the amino wart domain but lacked the carboxy hog domain. Together this made a family of ten warthog genes.
Previous research in my lab (the Woollard lab in the Biochemistry department at the University of Oxford) had linked the wrt-2 branch of the warthog family (figure 2) to defects in left right asymmetry, with wrt-2 and wrt-4 giving the highest penetrance phenotypes. However it was found that the GFP marker used in the previous strains to help quantify another phenotype (related to vulval development) caused defects of its own. This is where I come in. My project was to cross and create strains of worms without this GFP background and then requantify the left right asymmetry defects. I studied the single mutants wrt-2, wrt-4, wrt-8, as well as the double mutants wrt-4;wrt-2, wrt-8;wrt-2, wrt-8;wrt-4 and the triple mutant wrt-8;wrt-4;wrt-2. The double and triple mutant strains I created myself. The combined mutations allow investigation into redundancy between the genes, giving a glimpse into their evolutionary history. The wrt-7 gene has been shown to have no expression pattern and is likely a pseudogene, so due to my limited time in the lab I didn’t quantify the wrt-7 mutations.
On a side note: the C.elegans species is great. They’re heamaphrodites so you can just leave them on an agar plate to reproduce by themselves. Males do exist – enabling you to perform genetic crosses. They’re transparent, so no need to dissect anything. You can even freeze them and they’ll be alright in liquid nitrogen until someone needs them!
At first glance C.elegans may not seem very asymmetric. However one source of left right asymmetry is the relative positioning of the gonads and intestine either side of the vulva in the worm. This is best shown through pictures. In the wildtype worm (figure 3d) the picture in the lefthand plane shows that in the anterior part of the worm one can see intestinal cells and the intestinal lumen going down the centre. In the posterior part of the worm one can see the U shaped gonad that migrates away from the vulva (just visible in the bottom left hand corner of that picture) along the body, turns twice then travels back along the body (schematic figure 3c). The top picture of a whole worm (figure 3d) shows the opposite situation (the worm is laying on its other side) where the gonad can be seen at the anterior part of the body and the intestine at the posterior.
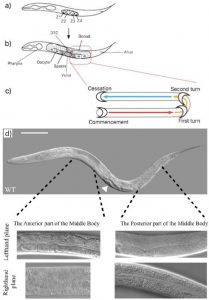
Thus the anterior/posterior distribution of gonad and intestine gives us a tool to study and quantify left right asymmetry in worms. After discussions with Emily (a recent biology graduate who had been working on the project before me) we decided to only quantify worms in which the posterior gonad was visible down the microscope. Thus, worms which I counted looked like those in the two lefthand plane pictures in figure 3d. This decision, although arbitrary in itself, allows our results when published to be more understandable and easier to replicate.
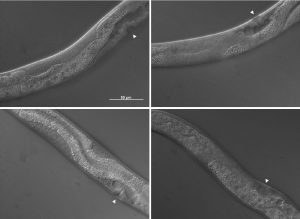
The pictures shown in figure 4 demonstrate some of the mutant phenotypes scored in this project. Basically I was looking for gonad tissue poking through anterior intestine tissue and intestine tissue poking through posterior gonad tissue. Worms with this phenotype were scored as mutant and were also further divided into anterior or posterior defects.
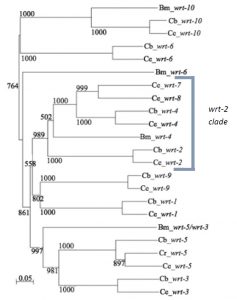
One of the most enjoyable parts of this project has been pipetting alongside Emily, who worked on this project before me and is continuing to develop it. In my 8 weeks I have seen first hand a small portion of the amount of research that has gone into studying the warthog genes. However chatting with Emily I have seen how my short project may in future count towards producing a scientific paper. At the moment Emily is taking the story of warthog genes in C.elegans back to its bioinformatics heart, producing a phylogenetic tree of all the warthog genes across the nematode family – trying to link evolution with phenotype. She is also expanding from the wrt-2 branch of the gene family into as many other members as she can get her hands on.
The highlight of my time in the project has been having so much independence in the lab, deciding day to day, week to week how I wanted to spend my time. This has been a fantastic insight into the life of a researcher and totally different to undergraduate labs. The lowlight would have to be the Saturday when I broke a pipette and in the process spilled a box of pipette tips all over my desk…
Thank you so much to everyone in the Woollard lab for being more friendly and welcoming than I could have hoped for.
Josie Elliott


 (6 votes)
(6 votes)