October in preprints
Posted by the Node, on 2 November 2017
Our latest monthly trawl for developmental biology (and other cool) preprints. Let us know if we missed anything.
October was a monster month for preprinting (a burst of post-summer productivity?), notable for the number of preprints covering plant development, disease modelling, modelling modelling, neurodevelopment, and organisms ranging from polychaete worms to hemipterans, Arctic charr to roses (plus all the usual suspects).
The preprints were hosted on bioRxiv, PeerJ, and arXiv. Use these links to get to the section you want:
Developmental biology
| Stem cells, regeneration & disease modelling
Evo-devo & evo
Cell biology
Modelling
Tools & resources
Research practice
Why not…
Developmental biology
| Patterning & signalling
Quantitative proteomic analysis of auxin signaling during seedling development. Dior Kelley, Zhouxin Shen, Justin W Walley, Elisabeth J Chapman, Steven P Briggs, Mark Estelle
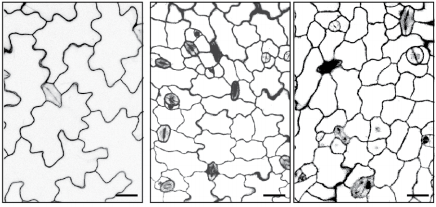
Lineage and stage-specific expressed CYCD7;1 coordinates the single symmetric division that creates stomatal guard cells. Annika K. Weimer, Juliana L. Matos, Walter Dewitte, Jim Murray, Dominique C. Bergmann
Stimulation of the final cell cycle in the stomatal lineage by the cyclin CYCD7;1 under regulation of the MYB transcription factor FOUR-LIPS. Farah Patell-Socha, David Newman, Eunkyoung Lee, Zidian Xie, Carl Collins, Erich Grotewold, James Murray, Walter Dewitte
The CLAVATA receptor FASCIATED EAR2 responds to different CLE peptides by signaling through different downstream effectors. Byoung Il Je, Fang Xu, Qingyu Wu, Lei Liu, Robert Meeley, David Jackson
Epidermal expression of a sterol biosynthesis gene regulates root growth by a non-cell autonomous mechanism in Arabidopsis. Eleri Short, Margaret Pullen, Gul Imriz, Dongbin Liu, Naomi Cope-Selby, Andrei Smertenko, Patrick J. Hussey, Jennifer F. Topping,Keith Lindsey
Postnatal Tendon Growth and Remodeling Requires Platelet-Derived Growth Factor Receptor Signaling. Kristoffer B. Sugg, James F. Markworth, Nathaniel P. Disser, Andrew M. Rizzi, Jeffrey R. Talarek, Dylan C. Sarver, Susan V. Brooks, Christopher L. Mendias
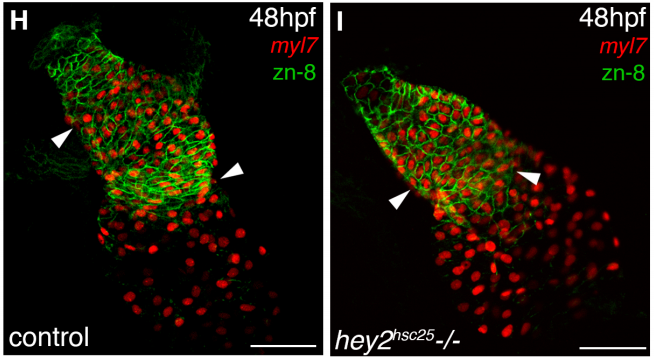
Hey2 restricts cardiac progenitor addition to the developing heart. Natalie Gibb, Savo Lazic, Ashish R. Deshwar, Xuefei Yuan, Michael D. Wilson, Ian C. Scott
ALKALs are in vivo ligands for ALK family Receptor Tyrosine Kinases in the neural crest and derived cells. Andrey Fadeev, Patricia Mendoza Garcia, Uwe Irion, Jikui Guan, Kathrin Pfeifer, Stephanie Wiessner, Fabrizio Serluca, Ajeet P Singh, Christiane Nusslein-Volhard, Ruth H. Palmer
Ttc21b is required in Bergmann glia during postnatal cerebellar development. Ashley M. Driver, Christopher Shumrick, Rolf Stottmann
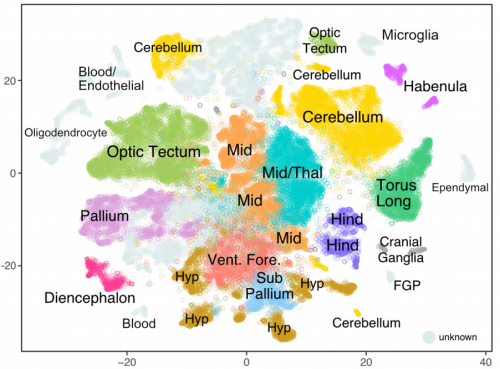
Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain by scGESTALT. Bushra Raj, Daniel E. Wagner, Aaron McKenna, Shristi Pandey, Allon M. Klein, Jay Shendure, James A. Gagnon, Alexander F. Schier
Massively parallel single cell lineage tracing using CRISPR/Cas9 induced genetic scars. Bastiaan Spanjaard, Bo Hu, Nina Mitic, Jan Philipp Junker
MYC activity is required for maintenance of the Neuromesodermal Progenitor signalling network and for correct timing of segmentation clock gene oscillations. Ioanna Mastromina, Laure Verrier, Kate G. Storey, J. Kim Dale
Coordination of robust single cell rhythms in the Arabidopsis circadian clock via spatial waves of gene expression. Peter D. Gould, Mirela Domijan, Mark Greenwood, Isao T. Tokuda, Hannah Rees, Laszlo Kozma-Bognar, Anthony J. Hall, James C. W. Locke
Transcription factor activity rhythms and tissue-specific chromatin interactions explain circadian gene expression across organs. Jake Yeung, Jerome Mermet, Celine Jouffe, Julien Marquis, Aline Charpagne, Frederic Gachon, Felix Naef
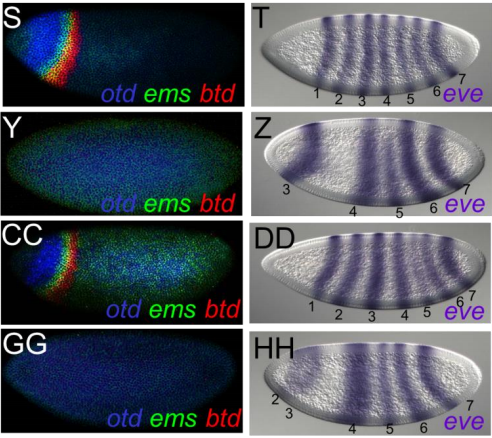
A Feed-forward Relay between Bicoid and Orthodenticle Regulates the Timing of Embryonic Patterning in Drosophila. Rhea Datta, Jia Ling, Jesse Kurland, Xiaotong Ren, Xu Zhe, Gozde Yucel, Jackie Moore, Leila Shokri, Isabel Baker, Timothy Bishop, Paolo Struffi, Rimma Levina, Martha Bulyk, Robert Johnston, Stephen Small
Proteolytic control of centrosome activity by APC/C-Polo maintains oocyte fate in Drosophila. Alexis L. Braun, Francesco Meghini, Gema Villa-Fombuena, Elisa M. Fernandez, Morgane Guermont, Maria D. Martin-Bermudo, Acaimo Gonzalez-Reyes, Yuu Kimata
spe-43 is required for sperm activation in C. elegans. Amber R Krauchunas, Ernesto Mendez, Julie Zhouli Ni, Marina Druzhinina, Amanda Mulia, Jean Parry, Sam Guoping Gu, Gillian M Stanfield, Andrew Singson
Planar Cell Polarity: What Does The prickle Gene Do? Jose Casal, Beatriz Ibanez-Jimenez, Peter A. Lawrence
Novel approach to quantitative spatial gene expression uncovers cryptic evolution in the developing Drosophila eye. Sammi Ali, Sarah Signor, Konstantin Kozlov, Sergey Nuzhdin
Csde1 cooperates with Strap to control translation of erythroid transcripts. Kat S. Moore, Nurcan Yagci, Floris van Alphen, Alexander B. Meijer, Peter A. C. ‘t Hoen, Marieke M. von Lindern
Transcriptional regulation by NR5A2 couples cell differentiation and inflammation in the pancreas. Isidoro Cobo, Paola Martinelli, Marta Flandez, Latifa Bakiri, Mingfeng Zhang, Enrique Carrillo-de-Santa-Pau, Jinping Jia, Liv Thommesen, Torunn Bruland, Natalia del Pozo, Sara Olson, Jill Smith, William R. Bamlet, Gloria M. Petersen, Nuria Malats, Laufey Amundadottir, Erwin Wagner, Francisco X. Real
RhoGEF9 splice isoforms influence neuronal maturation and synapse formation downstream of a2 GABAA receptors. Claire deGroot, Amalia Floriou-Servou, Yuan-Chen Tsai, Simon Frueh, Manuela Kohler, Georgia Parkin, Cornelia Schwerdel, Giovanna Bosshard, Kai Kaila, Jean-Marc Fritschy, Shiva Tyagarajan
Microglial elimination of dopamine D1 receptors defines sex-specific changes in nucleus accumbens development and social play behavior during adolescence. Ashley Kopec, Caroline J. Smith, Nathan R. Ayre, Sean C. Sweat, Staci D. Bilbo
Experience-dependent neuroplasticity of the developing hypothalamus: integrative epigenomic approaches. Annie Vogel Ciernia, Ben I Laufer, Keith W Dunaway, Charles E Mordaunt, Rochelle L Coulson, Theresa S Totah, Danielle S Stolzenberg, Jaime C Frahm, Akanksha Singh-Taylor, Tallie Z Baram, Janine M LaSalle, Dag H Yasui
Mast Cells in the Developing Brain Determine Adult Sexual Behavior. Kathryn M. Lenz, Lindsay A. Pickett, Christopher L. Wright, Katherine T. Davis, Anabel Galan, Margaret M. McCarthy
Development differentially sculpts receptive fields across human visual cortex. Jesse Gomez, Vaidehi Natu, Brianna Jeska, Michael Barnett, Kalanit Grill-Spector
Cortical Connectivity In A Macaque Model Of Congenital Blindness. Loic Magrou, Pascal Barone, Nikola T. Markov, Herbert Killackey, Pascale Giroud, Michel Berland, Kenneth Knoblauch, Colette Dehay, Henry Kennedy
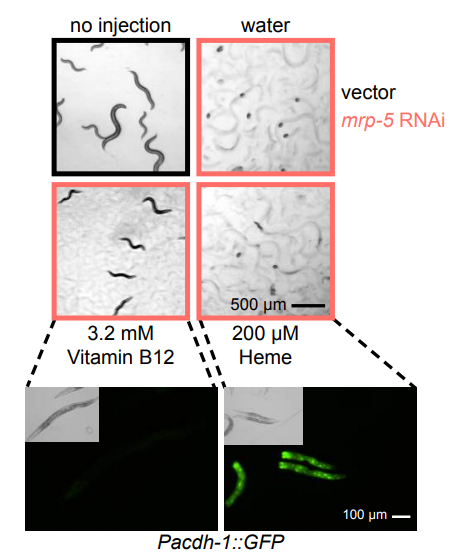
C. elegans Multidrug Resistance Protein 5 (MRP-5) Transports Vitamin B12 from the Intestine to the Gonad to Support Embryonic Development. Huimin Na, Olga Ponomarova, Gabrielle E. Giese, Marian J. M. Walhout
Hub connectivity, neuronal diversity, and gene expression in the C. elegans connectome. Aurina Arnatkeviciute, Ben D. Fulcher, Roger Pocock, Alex Fornito
The Emergent Connectome in Caenorhabditis elegans Embryogenesis. DevoWorm Group, Bradly J. Alicea
| Morphogenesis & mechanics
Mechanical stretching can modify the axial morphology of early chick embryos. Ben K. A. Nelemans, Manuel Schmitz, Hannan Tahir, Roeland M. H. Merks, Theodoor H. Smit
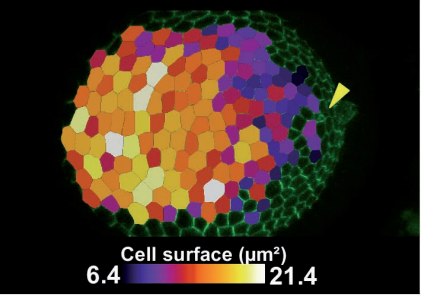
Jak-Stat pathway induces Drosophila follicle elongation by a gradient of apical contractility. Herve Alegot, Pierre Pouchin, Olivier Bardot, Vincent Mirouse
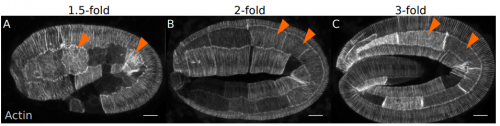
Coordinated morphogenesis through tension-induced planar polarity. Ghislain Gillard, Ophelie Nicolle, Thibault Brugiere, Sylvain Prigent, Mathieu Pinot, Gregoire Michaux
Zebrafish yolk syncytial nuclei migrate along a dynamic microtubule network. Zhonghui Fei, Koeun Bae, Serge Parent, Katharine Goodwin, Guy Tanentzapf, Ashley Bruce
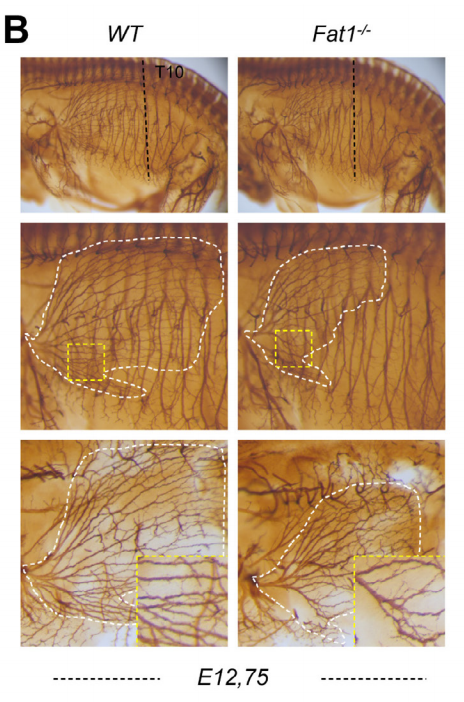
Tissue-specific activities of the Fat1 cadherin cooperate to control neuromuscular morphogenesis. Francoise Helmbacher
Development of migrating entheses involves replacement of progenitor populations. Neta Felsenthal, Sarah Rubin, Tomer Stern, Sharon Krief, Deepanwita Pal, Brian A. Pryce, Ronen Schweitzer, Elazar Zelzer
Requirement of the Dynein-adaptor Spindly for mitotic and post-mitotic functions in Drosophila. Giuliana D. Clemente, Matthew R. Hannaford, Jens Januschke, Eric R. Griffis, Hans-Arno J. Muller
MAK and CCRK kinases regulate kinesin-2 motility in C. elegans neuronal cilia. Peishan Yi, Chao Xie, Guangshuo Ou
Bundle sheath extensions in tomato affect leaf phenotypic plasticity in response to irradiance. Maria Antonia Machado Barbosa, Daniel H. Chitwood, Aristéa A. Azevedo, Wagner L. Araújo, Samuel C. V. Martins, Dimas M. Ribeiro, Lázaro E. P. Peres, Agustin Zsögön
Dynamics of pollen tube growth and embryo sac development in Pozna Plava plum cultivar related to fruit set. Milena Đorđević, Radosav Cerović, Sanja Radičević, Dragan Nikolić, Nebojša Milošević, Ivana Glišić, Slađana Marić, Milan Lukić
Bimodal Spindle Orientation Drives Tissue Regularity in a Proliferating Epithelium. Tara M. Finegan, Daxiang Na, Austin V. Skeeters, Nicole S. Dawney, Patrick W. Oakes, Alexander G. Fletcher, Dan T. Bergstralh
| Genes & genomes
Single-cell transcriptional regulations and accessible chromatin landscape of cell fate decisions in early heart development. Guangshuai Jia, Jens Preussner, Stefan Guenther, Xuejun Yuan, Michail Yekelchyk, Carsten Kuenne, Mario Looso, Yonggang Zhou, Thomas Braun
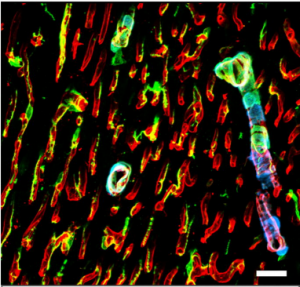
Single cell transcriptional profiling reveals cellular diversity, communication, and sexual dimorphism in the mouse heart. Daniel A. Skelly, Galen T. Squiers, Micheal A. McLellan, Mohan T. Bolisetty, Paul Robson, Nadia Rosenthal, Alexander R. Pinto
Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. Ralf Gilsbach, Martin Schwaderer, Sebastian Preissl, Bjoern A Gruening, David Kranzhoefer, Pedro Schneider, Thomas G Nuehrenberg, Sonia Mulero-Navarro, Dieter Weichenhan, Christian Braun, Martina Dressen, Adam R Jacobs, Harald Lahm, Torsten Doenst, Rolf Backofen, Markus Krane, Bruce D Gelb, Lutz Hein
Single-cell transcriptomic catalog of mouse cortical development. Lipin Loo, Jeremy M Simon, Eric S McCoy, Jesse K Niehaus, Mark J Zylka
Comprehensive single cell RNAseq analysis of the kidney reveals novel cell types and unexpected cell plasticity. Jihwan Park, Rojesh Shrestha, Chengxiang Qiu, Ayano Kondo, Shizheng Huang, Max Werth, Mingyao Li, Jonathan Barasch, Katalin Susztak
The Transcriptional Logic of Mammalian Neuronal Diversity. Ken Sugino, Erin Clark, Anton Schulmann, Yasuyuki Shima, Lihua Wang, David L. Hunt, Bryan M. Hooks, Dimitri Trankner, Jayaram Chandrashekar, Serge Picard, Andrew Lemire, Nelson Spruston, Adam Hantman, Sacha B. Nelson
RNA velocity in single cells. Gioele La Manno, Ruslan Soldatov, Hannah Hochgerner, Amit Zeisel, Viktor Petukhov, Maria Kastriti, Peter Lonnerberg, Alessandro Furlan, Jean Fan, Zehua Liu, David van Bruggen, Jimin Guo, Erik Sundstrom, Goncalo Castelo-Branco, Igor Adameyko, Sten Linnarsson, Peter Kharchenko
Unbiased association and expression studies identify novel genes for tooth development. Meredith Williams, Claudia Biguetti, Miguel Romero-Bustillos, Kanwal Maheshwari, Nuriye Dinckan, Franco Cavalla, Xiaoming Liu, Renato Silva, Sercan Akyalcin, Z O Uyguner, Alexandre Vieira, Brad A Amendt, Walid D Fakhrouri, Ariadne Letra
Structural robustness of mammalian transcription factor networks reveals plasticity across development. Jose Luis Caldu-Primo, Elena Alvarez-Buylla, Jose Davila-Velderrain
Sexual lineage specific DNA methylation regulates Arabidopsis meiosis. James Walker, Hongbo Gao, Jingyi Zhang, Billy Aldridge, Martin Vickers, James D Higgins, Xiaoqi Feng
Placeholder nucleosomes underlie germline-to-embryo DNA methylation reprogramming. Patrick J. Murphy, ShanFu Wu, Cody R. James, Candice L. Wike, Bradley R. Cairns
Hypermethylation of human DNA: Fine-tuning transcription associated with development. Carl Baribault, Kenneth C. Ehrlich, V. K. Chaithanya Ponnaluri, Sriharsa Pradhan, Michelle Lacey, Melanie Ehrlich
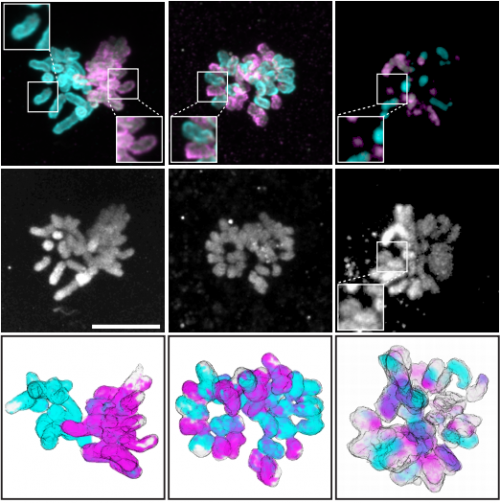
Disruption of dual zygotic spindle assembly shows epigenetic asymmetry to be chromosome intrinsic. Judith Reichmann, Bianca Nijmeijer, M. Julius Hossain, Manuel Eguren, Isabell Schneider, Antonio Z. Politi, M. Julia Roberti, Lars Hufnagel, Takashi Hiiragi, Jan Ellenberg
Two coupled feedback loops explain random mono-allelic Xist upregulation at the onset of X-chromosome inactivation. Verena Mutzel, Ikuhiro Okamoto, Ilona Dunkel, Mitinori Saitou, Luca Giorgetti, Edith Heard, Edda G. Schulz
Paternal easiRNAs regulate parental genome dosage in Arabidopsis. German Martinez, Philip Wolff, Zhenxing Wang, Jordi Moreno-Romero, Juan Santos-Gonzalez, Lei Liu Conze, Christopher DeFraia, Keith Slotkin, Claudia Kohler
microRNA-triggered transposon small RNAs mediate genome dosage response. Filipe Borges, Jean-Sebastien Parent, Frederic van Ex, Philip Wolff, German Martinez, Claudia Kohler, Robert A Martienssen
LIN28 selectively modulates a subclass of let-7 microRNAs. Dmytro Ustianenko, Hua-Sheng Chiu, Sebastien M. Weyn-Vanhentenryck, Pavel Sumazin, Chaolin Zhang
Preovulatory oocyte aging in mice affects fertilization rate and embryonic genome activation. Hannah Demond, Debora Dankert, Ruth Gruemmer, Bernhard Horsthemke
Universal transcriptomic signature of age reveals temporal scaling of Caenorhabditis elegans aging trajectories. Andrei E. Tarkhov, Ramani Alla, Srinivas Ayyadevara, Mikhail Pyatnitskiy, Leonid Menshikov, Robert Shmookler Reis, Peter O. Fedichev
The Drosophila Eukaryotic Initiation Factor eIF6 affects development by regulating apoptosis via the ecdysone pathway. Arianna Russo, Guido Gatti, Roberta Alfieri, Elisa Pesce, Kelly Soanes, Sara Ricciardi, Cristina Cheroni, Thomas Vaccari, Stefano Biffo, Piera Calamita
L(3)mbt and the LINT complex safeguard tissue identity in the Drosophila ovary. Remi-Xavier Coux, Felipe Karam Teixeira, Ruth Lehmann
Cooperative recruitment of Yan to paired high affinity ETS sites organizes repression to confer specificity and robustness to cardiac cell fate specification. Jean-Francois Boisclair Lachance, Jemma L. Webber, Ilaria Rebay
A conserved maternal-specific repressive domain in Zelda revealed by Cas9-mediated mutagenesis in Drosophila melanogaster. Danielle C. Hamm, Elizabeth D. Larson, Markus Nevil, Kelsey E. Marshall, Eliana R. Bondra, Melissa M. Harrison
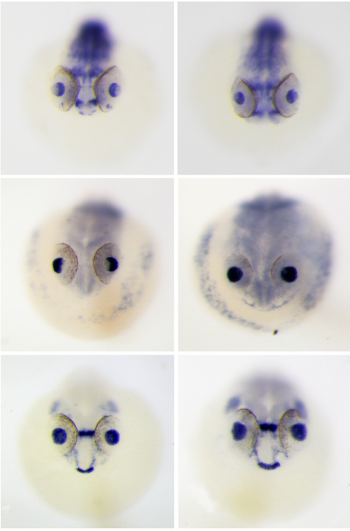
The DEAD box RNA helicase Ddx39a is essential for myocyte and lens development in zebrafish. Linlin Zhang, Beibei Li, Yuxi Yang, Ian C. Scott, Xin Lou
RES complex is associated with intron definition and required for zebrafish early embryogenesis. Juan Fernandez, Miguel Moreno-Mateos, Andre Gohr, Shun Chan, Manuel Irimia, Antonio Giraldez
An evolutionarily conserved leucine zipper-like motif accounts for strong tetramerization capabilities of SEPALLATA-like MADS-domain transcription factors controlling flower development. Florian Ruempler, Guenter Theissen, Rainer Melzer
Massive variation of short tandem repeats with functional consequences across strains of Arabidopsis thaliana. Maximilian O. Press, Rajiv C. McCoy, Ashley N. Hall, Joshua M. Akey, Christine Queitsch
Transcriptome dynamics at the Arabidopsis graft junction reveal an inter-tissue recognition mechanism that activates vascular regeneration. Charles W. Melnyk, Alexander Gabel, Thomas J. Hardcastle, Sarah Robinson, Shunsuke Miyashima, Ivo Grosse, Elliot M. Meyerowitz
A study of allelic series using transcriptomic phenotypes in a metazoan. David Angeles-Albores, Paul Sternberg
Lamins organize the global three-dimensional genome from the nuclear periphery. Xiaobin Zheng, Jiabiao Hu, Sibiao Yue, Lidya Kristiani, Miri Kim, Michael Sauria, James Taylor, Youngjo Kim, Yixian Zheng
Single-cell absolute contact probability detection reveals that chromosomes are organized by multiple, low-frequency yet specific interactions. Diego I. I. Cattoni, Andres M. Cardozo Gizzi, Mariya Georgieva, Marco Di Stefano, Alessandro Valeri, Delphine Chamousset, Christophe Houbron, Stephanie Dejardin, Jean-Bernard Fiche, Inma Gonzalez, Jia-Ming Chang, Thomas Sexton, Marc A. Marti-Renom, Frederic Bantignies, Giacomo Cavalli, Marcelo Nollmann
Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Andrew Daniel Stephens, Patrick Z. Liu, Edward J. Banigan, Luay M Almassalha, Vadim Backman, Stephen A. Adam, Robert Goldman, John Marko
Distinct stimulatory mechanisms regulate the catalytic activity of Polycomb Repressive Complex 2 (PRC2). Chul-Hwan Lee, Marlene Holder, Daniel Grau, Ricardo Saldana-Meyer, Rais Ahmad Ganai, Jenny Zhang, Miao Wang, Marc-Werner Dobenecker, Danny Reinberg, Karim-Jean Armache
Distinct Epigenomic Patterns Are Associated with Haploinsufficiency and Predict Risk Genes of Developmental Disorders. Xinwei Han, Siying Chen, Elise D. Flynn, Dana Wintner, Yufeng Shen
The kinetics of pre-mRNA splicing in the Drosophila genome: influence of gene architecture. Athma A Pai, Telmo Henriques, Kayla McCue, Adam Burkholder, Karen Adelman, Christopher B Burge
Transcription start site profiling uncovers divergent transcription and enhancer-associated RNAs in Drosophila melanogaster. Michael P. Meers, Karen Adelman, Robert J. Duronio, Brian D. Strahl, Daniel J. McKay, A. Gregory Matera
Transgenerational dynamics of rDNA copy number in Drosophila male germline stem cells. Kevin L. Lu, Jonathan O. Nelson, Natalie Warsinger-Pepe, Yukiko M. Yamashita
Autosomal recessive coding variants explain only a small proportion of undiagnosed developmental disorders in the British Isles.Hilary C. Martin, Wendy D. Jones, James Stephenson, Juliet Handsaker, Giuseppe Gallone, Jeremy F. McRae, Elena Prigmore, Patrick Short, Mari Niemi, Joanna Kaplanis, Elizabeth Radford, Nadia Akawi, Meena Balasubramanian, John Dean, Rachel Horton, Alice Hulbert, Diana S. Johnson, Katie Johnson, Dhavendra Kumar, Sally Ann Lynch, Sarju G. Mehta, Jenny Morton, Michael J. Parker, Miranda Splitt, Peter D. Turnpenny, Pradeep C. Vasudevan, Michael Wright, Caroline F. Wright, David R. FitzPatrick, Helen V. Firth, Matthew E. Hurles, Jeffrey C. Barrett
| Stem cells, regeneration & disease modelling
Pattern analysis of pluripotency reveals the molecular basis of naïve, primed and formative states. Patrick S. Stumpf, Ben D. MacArthur
A common molecular logic determines embryonic stem cell self-renewal and reprogramming. Sara-Jane Dunn, Meng Amy Li, Elena Carbognin, Austin G Smith, Graziano Martello
Morphogen dynamics control patterning in a stem cell model of the human embryo. Idse Heemskerk, Kari Burt, Matthew Miller, Sapna Chabra, M. Cecilia Guerra, Aryeh Warmflash
Human pluripotent reprogramming with CRISPR activators. Jere Weltner, Diego Balboa, Shintaro Katayama, Maxim M. Bespalov, Kaarel Krjutskov, Eeva-Mari Jouhilahti, Ras Trokovic, Juha Kere, Timo Otonkoski
Subtle Changes in Clonal Dynamics Underlie the Age-Related Decline in Neurogenesis. Lisa Bast, Filippo Calzolari, Michael Strasser, Jan Hasenauer, Fabian Theis, Jovica Ninkovic, Carsten Marr
Conservation of differentiation but transformation of initiation in hippocampal neurogenesis. Hannah Hochgerner, Amit Zeisel, Peter Lönnerberg, Sten Linnarsson
Adult Neurogenesis Conserves Hippocampal Learning Capacity. Jahangir Alam, Takashi Kitamura, Yoshito Saitoh, Noriaki Ohkawa, Takashi Kondo, Kaoru Inokuchi
A de novo missense mutation in TUBA1A results in reduced neural progenitor survival and differentiation. Ashley Driver, Amy Pitstick, Chris Mayhew, Beth Kline-Fath, Howard Saal, Rolf Stottmann
Cftr Modulates Wnt/β-Catenin Signaling and Stem Cell Proliferation in Murine Intestine. Ashlee M. Strubberg, Jinghua Liu, Nancy M. Walker, Casey D. Stefanski, R. John MacLeod, Scott T. Magness, Lane L. Clarke
Division-independent differentiation mandates proliferative competition among stem cells. Amy Reilein, David Melamed, Simon Tavare, Daniel Kalderon
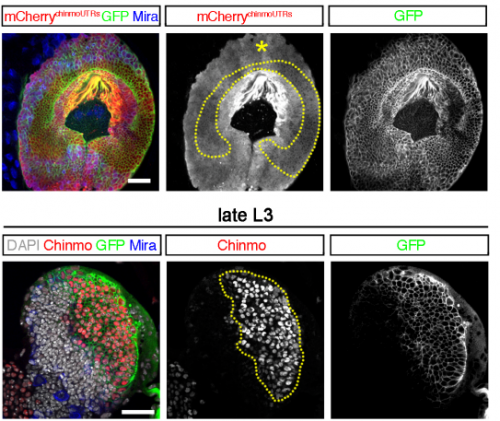
Two distinct mechanisms silence chinmo in Drosophila neuroblasts and neuroepithelial cells to limit their self-renewal. Caroline Dillard, Karine Narbonne-Reveau, Sophie Foppolo, Elodie Lanet, Cedric Maurange
p53 is required for female germline stem cell maintenance in P-element hybrid dysgenesis. Sadia Tasnim, Erin S. Kelleher
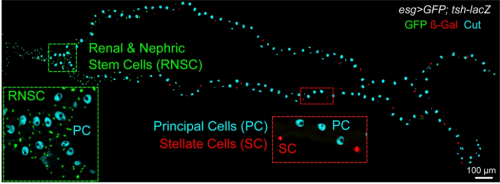
Shavenbaby and Yorkie mediate Hippo signaling to protect adult stem cells from apoptosis. Jerome Bohere, Alexandra Mancheno-Ferris, Kohsuke Akino, Yuya Yamabe, Sachi Inagaki, Helene Chanut-Delalande, Serge Plaza, Yuji Kageyama, Dani Osman, Cedric Polesello, Francois Payre
Comparative Analysis of Gene Expression Identifies Distinct Molecular Signatures of Bone Marrow- and Periosteal-Skeletal Stem/Progenitor Cells. Lorenzo Deveza, Kevin Lei, Laura Ortinau, Dongsu Park
Csde1 binds transcripts involved in protein homeostasis and controls their expression in erythropoiesis. Kat S. Moore, Nurcan Yagci, Floris van Alphen, Nahuel A. Paolini, Rastislav horos, Ntsiki M. Held, Riekelt H. Houtkooper, Emile van den Akker, Alexander B. Meijer, Peter A. C. ‘t Hoen, Marieke von Lindern
Nitrate modulates stem cell dynamics by regulating WUSCHEL expression through cytokinins. Benoit Landrein, Pau Formosa-Jordan, Alice Malivert, Christoph Schuster, Charles W. Melnyk, Weibing Yang, Colin Turnbull, Elliot M. Meyerowitz, James C.W. Locke, Henrik Jonsson
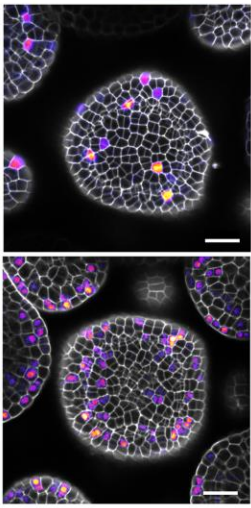
Cell Cycle Control by Nuclear Sequestration of CDC20 and CDH1 mRNA in Plant Stem Cells. Weibing Yang, Raymond Wightman, Elliot M. Meyerowitz
Effects of Ivermectin exposure on regeneration of D. dorotocephala planaria. Nina N. Ferenc, Michael Levin
Temporal differentiation of bovine airway epithelial cells grown at an air-liquid interface. Daniel Cozens, Erin Sutherland, Francesco Marchesi, Geraldine Taylor, Catherine C. Berry, Robert L. Davies
Bacterial Colonization Stimulates A Complex Physiological Response In The Immature Human Intestinal Epithelium. David R. Hill, Sha Huang, Melinda S. Nagy, Veda K. Yadagiri, Courtney Fields, Dishari Mukherjee, Brooke Bons, Priya H. Dedhia, Alana M. Chin, Yu-Hwai Tsai, Shrikar Thodla, Thomas M. Schmidt, Seth Walk, Vincent B. Young, Jason R. Spence
In vitro induction and in vivo engraftment of lung bud tip progenitor cells derived from human pluripotent stem cells. Alyssa J. Miller, David R. Hill, Melinda S. Nagy, Yoshiro Aoki, Briana R. Dye, Alana M. Chin, Sha Huang, Felix Zhu, Eric S. White, Vibha Lama, Jason R. Spence
Identification, Isolation, and Characterization of Human LGR5-positive Colon Adenoma Cells. Michael K Dame, Durga Attili, Shannon D McClintock, Priya H Dedhia, Peter Ouilette, Olaf Hardt, Alana M Chin, Xiang Xue, Julie Laliberte, Erica L Katz, Gina M Newsome, David Hill, Alyssa Miller, Yu-Hwai Tsai, David Agorku, Christopher H Altheim, Andreas Bosio, Becky Simon, Linda C Samuelson, Jay A Stoerker, Henry D Appelman, James Varani, Max S Wicha, Dean E Brenner, Yatrik M Shah, Jason R Spence, Justin A Colacino
Single-cell transcriptome analysis identifies distinct cell types and intercellular niche signaling in a primary gastric organoid model. Jiamin Chen, Billy T. Lau, Noemi Andor, Sue M. Grimes, Christine Handy, Christina Wood-Bouwens, Hanlee P. Ji
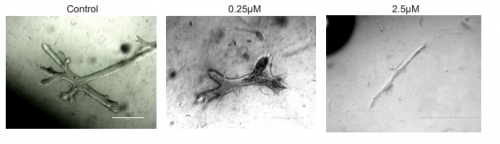
Cadmium exposure inhibits branching morphogenesis and causes alterations consistent with HIF-1α inhibition in human primary breast organoids. Sabrina A. Rocco, Lada Koneva, Lauren Y. M. Middleton, Tasha Thong, Sumeet Solanki, Sarah Karram, Kowit Nambunmee, Craig Harris, Laura S. Rozek, Maureen A. Sartor, Yatrik M. Shah, Justin Colacino
Gene expression in patient-derived neural progenitors provide insights into neurodevelopmental aspects of schizophrenia. Oleg V Evgrafov, Chris Armoskus, Bozena B Wrobel, Valeria N Spitsyna, Tade Souaiaia, Jenifer S Herstein, Christopher P Walker, Joseph D Nguyen, Adrian Camarena, Jonathan R Weitz, Jae Mun ‘Hugo’ Kim, Edder Lopez Duarte, Kai Wang, George M Simpson, Janet L Sobell, Helena Medeiros, Michele T Pato, Carlos N Pato, James A Knowles
Transcriptomic Dynamics of a non-coding trinucleotide repeat expansion disorder SCA12 in iPSC derived neuronal cells: signatures of interferon induced response. Deepak Kumar, Parashar Dhapola, Ashaq Hussain, Rintu Kutum, Achal Kumar Srivastava, Mitali Mukerji, Odity Mukherjee, Mohammed Faruq
The Polycomb-dependent epigenome controls β-cell dysfunction, dedifferentiation and diabetes. Tess Tsai-Hsiu Lu, Steffen Heyne, Erez Dror, Eduard Casas, Laura Leonhardt, Thorina Boenke, Chih-Hsiang Yang, Sagar, Laura Arrigoni, Kevin Dalgaard, Raffaele Teperino, Lennart Enders, Madhan Selvaraj, Marius Ruf, Sunil Jayaramaiah Raja, Huafeng Xie, Ulrike Boenisch, Stuart H. Orkin, Francis Lynn, Brad G. Hoffman, Dominic Grün, Tanya Vavouri, Adelheid Lempradl, Andrew Pospisilik
Whole Genomes Define Concordance of Matched Primary, Xenograft, and Organoid Models of Pancreas Cancer. Deena M.A. Gendoo, Robert E. Denroche, Amy Zhang, Gun Ho Jang, Mathieu Lemire, Sandra Fischer, Nikolina Radulovich, Dianne Chadwick, Ilinca M. Lungu, Lincoln D. Stein, Julie M. Wilson, John M.S. Bartlett, Ming-Sound Tsao, Neesha Dhani, David Hedley, Steven Gallinger, Benjamin Haibe-Kains
A novel zebrafish intestinal tumor model reveals a role for cyp7a1-dependent tumor-liver crosstalk in tumor’s adverse effects on host. Sora Enya, Koichi Kawakami, Yutaka Suzuki, Shinpei Kawaoka
highroad is induced by retinoids and clears mutant Rhodopsin-1 in Drosophila Retinitis Pigmentosa models. Huai-Wei Huang, Brian Brown, Jaehoon Chung, Pedro M. Domingos, Hyung Don Ryoo
The BXD Mouse Strains are a Model System for Studying Optic Nerve Regeneration. Jiaxing Wang, Ying Li, Rebecca King, Felix L. Struebing, Eldon E. Geisert
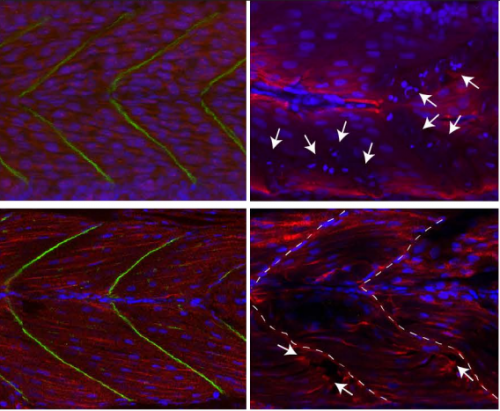
Epigenetic Regulators Modulate Muscle Damage In Duchenne Muscular Dystrophy Model. Fernanda Bajanca, Laurence Vandel
Interleukins 4 and 13 Induce Exon Skipping of Mutant Dystrophin Pre-mRNA to Restore Dystrophin Production. SiewHui Low, Chen-Ming Fan
Galectin-3 and N-acetylglucosamine promote myogenesis and improve skeletal muscle function in the mdx model of Duchenne muscular dystrophy. Ann Rancourt, Sebastien Dufresne, Guillaume St-Pierre, Julie-Christine Levesque, Haruka Nakamura, Yodai Kikuchi, Masahiko S. Satoh, Jerome Frenette, Sachiko Sato
Ift172 conditional knockout mice exhibit rapid retinal degeneration and protein trafficking defects. Priya R. Gupta, Nachiket Pendse, Scott H. Greenwald, Mihoko Leon, Qin Liu, Eric A. Pierce, Kinga Maria Bujakowska
Genomic Locus Modulating IOP in the BXD RI Mouse Strains. Rebecca A. King, Ying Li, Jiaxing Wang, Felix Struebing, Eldon E. Geisert
WDR11-mediated Hedgehog signalling defects underlie a new ciliopathy related to Kallmann syndrome. Yeon-Joo Kim, Daniel Osborn, Ji-Young Lee, Masatake Araki, Kimi Araki, Timothy Mohun, Johanna E M Kansakoski, Nina Brandstack, Hyun-Taek Kim, Francesc Miralles, Cheol-Hee Kim, Nigel A Brown, Hyung-Goo Kim, Juan Pedro Martinez-Barbera, Paris Ataliotis, Taneli Raivio, Lawrence C Layman, Soo-Hyun Kim
Automated microfluidic cell culture of stem cell derived dopaminergic neurons in Parkinson’s disease. Khalid I. W. Kane, Edinson Lucumi Moreno, Siham Hachi, Moriz Walter, Javier Jarazo, Thomas Hankemeier, Paul Vulto, Jens Schwamborn, Martin Thoma, Ronan M. T. Fleming
Mutations in C. elegans neuroligin-like glit-1, the apoptosis pathway and the calcium chaperone crt-1 increase dopaminergic neurodegeneration after 6-OHDA treatment. Sarah-Lena Offenburger, Elisabeth Jongsma, Anton Gartner
C11orf70 mutations causing primary ciliary dyskinesia disrupt a conserved step in the intraflagellar transport-dependent assembly of multiple axonemal dyneins. Mahmoud R. Fassad, Amelia Shoemark, Pierrick le Borgne, France Koll, Mitali Patel, Mellisa Dixon, Jane Hayward, Charlotte Richardson, Emily Frost, Lucy Jenkins, Thomas Cullup, Eddie M. K. Chung, Michel Lemullois, Anne Aubusson-Fleury, Claire Hogg, David R. Mitchell, Anne-Marie Tassin, Hannah M. Mitchison
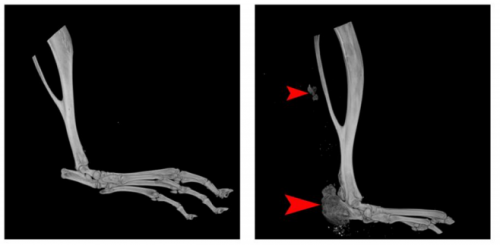
Characterization of a Novel Model of Overuse-Induced Calcific Achilles Tendinopathy in Mice: Contralateral Tendinopathy Following Unilateral Tenotomy. Wang Liang, Zhang Jie, Yin Gang-hui, Zhang Zhong-min, Chen Tian-yu, Jin Jian, Lai Ping-lin, Huang Bin, Yan Bo, Chen Yu-hui, Jin Da-di, Huang Min-jun
Osteo-Oto-Hepato-Enteric Syndrome (O2HE) is caused by loss of function mutations in UNC45A. Clothilde Esteve, Ludmila Francescatto, Perciliz Tan, Aurelie Bourchany, Cecile De Leusse, Evelyne Marinier, Arnaud Blanchard, Patrice Bourgeois, Celine Brochier-Armanet, Ange-Line Bruel, Arnauld Delarue, Yannis Duffourd, Emmanuelle Ecochard-Dugelay, Philippe Gauchez, Geraldine Hery, Frederic Huet, Philippe Gauchez, Emmanuel Gonzales, Catherine Guettier-Bouttier, Mina Komuta, Caroline Lacoste, Raphaelle Maudinas, Karin Mazodier, Yves Rimet, Jean-Baptiste Riviere, Bertrand Roquelaure, Sabine Sigaudy, Xavier Stephenne, Christel Thauvin-Robinet, Julien Thevenon, Jacques Sarles, Nicolas Levy, Catherine Badens, Olivier Goulet, Jean-Pierre Hugot, Nicholas Katsanis, Laurence Faivre, Alexandre Fabre
Contribution of structural and intronic mutations to RPGRIP1-mediated inherited retinal dystrophies. Farzad Jamshidi, Emily M Place, Daniel Navarro-Gomez, Mathew Maher, Elise Valkanas, Monkol Lek, Daniel MacArthur, Kinga M Bujakowska, Eric A Pierce
Targeted deletion of Nmnat1 in mouse retina leads to early severe retinal dystrophy. Xiaolin Wang, Yu Fang, Rongsheng Liao, Tao Wang
Protective effect of Rhus coriaria fruit extracts against hydrogen peroxide-induced oxidative stress in muscle progenitors and zebrafish embryos. Fadia Najjar1, Francine Rizk1, Gilles Carnac2, Rim Nassar1, Sara Jabak1, Anatoly Petrovich Sobolev3, Yara Bou Saada4, Marwan El Sabban5, Aline Hamade1
Evo-devo & evo
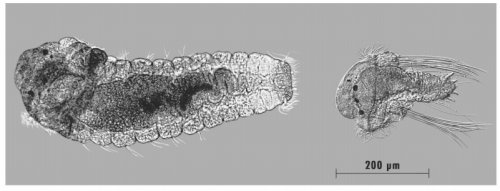
Decoupled Maternal and Zygotic Genetic Effects Shape the Evolution of Development. Christina Zakas, Jennifer Deutscher, Alex Kay, Matthew Rockman
Hox-logic of preadaptations for social insect symbiosis in rove beetles. Joseph Parker, K. Taro Eldredge, Isaiah Thomas, Rory Coleman, Steven Davis
Cis-regulatory evolution integrated the Bric-à-brac transcription factors into a novel fruit fly gene regulatory network. Thomas Michael Williams, Mark Rebeiz, Eric M Camino, Maxwell J Roeske, Sumant Grover
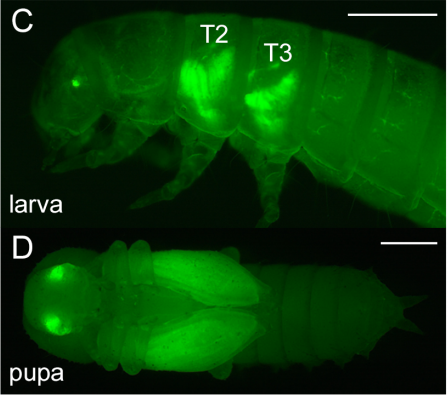
Enhancer identification and activity evaluation in the red flour beetle, Tribolium castaneum. Yi-Ting Lai, Kevin D. Deem, Ferran Borras-Castells, Nagraj Sambrani, Heike Rudolf, Kushal Suryamohan, Ezzat El-Sherif, Marc S. Halfon, Daniel J. McKay, Yoshinori Tomoyasu
Gender- and gamete-specific patterns of X chromosome segregation in a three-gendered nematode. Sophie Tandonnet, Maureen C. Farrell, Georgios Koutsovoulos, Mark Blaxter, Manish Parihar, Penny L. Sadler, Diane Shakes, Andre Pires da Silva
An epigenetic mechanism for cavefish eye degeneration. Aniket Gore, Kelly Tomins, James Iben, Li Ma, Daniel Castranova, Andrew Davis, Amy Parkhurst, William Jeffery, Brant Weinstein
Evidence for Late Pleistocene origin of Astyanax mexicanus cavefish. Julien Fumey, Hélène Hinaux, Céline Noirot, Claude Thermes, Sylvie Rétaux, Didier Casane
Identifying developmentally important genes with single-cell RNA-seq from an embryo. Garth R Ilsley, Ritsuko Suyama, Takeshi Noda, Nori Satoh, Nicholas M Luscombe

Differential gene expression during early development in recently evolved and sympatric Arctic charr morphs. Jóhannes Guðbrandsson, Sigríður Rut Franzdóttir, Bjarni Kristófer Kristjánsson, Ehsan Pashay Ahi, Valerie Helene Maier, Sigurður Sveinn Snorrason, Zophonías Oddur Jónsson, Arnar Pálsson
Placozoans are eumetazoans related to Cnidaria. Christopher E. Laumer, Harald Gruber-Vodicka, Michael G. Hadfield, Vicki B. Pearse, Ana Riesgo, John C. Marioni, Gonzalo Giribet
Lessons from simple marine models on the bacterial regulation of eukaryotic development. Arielle Woznica, Nicole King
Motility in multicellular Chlamydomonas reinhardtii. Margrethe Boyd, Matthew D Herron, Frank Rosenzweig
Microinjection to deliver protein, mRNA, and DNA into zygotes of the cnidarian endosymbiosis model Aiptasia sp. Victor Arnold Shivas Jones, Madeline Bucher, Elizabeth Ann Hambleton, Annika Guse
Reproductive mode, stem cells and regeneration in a freshwater cnidarian with post-reproductive senescence. Flora Sebestyen, Zoltan Barta, Jacint Tokolyi
Cnidarian cell type diversity revealed by whole-organism single-cell RNA-seq analysis. Arnau Sebe-Pedros, Elad Chomsky, Baptiste Saudemont, Marie-Pierre Mailhe, Flora Pleisser, Justine Renno, Yann Loe-Mie, Aviezer Lifshitz, Zohar Mukamel, Sandrine Schmutz, Sophie Nouvault, Francois Spitz, Amos Tanay, Heather Marlow

Molecular evolutionary trends and feeding ecology diversification in the Hemiptera, anchored by the milkweed bug genome. Kristen A. Panfilio, Iris M. Vargas Jentzsch, Joshua B. Benoit, Deniz Erezyilmaz, Yuichiro Suzuki, Stefano Colella, Hugh M. Robertson, Monica F. Poelchau, Robert M. Waterhouse, Panagiotis Ioannidis, Matthew T. Weirauch, Daniel S.T. Hughes, Shwetha C. Murali, John H. Werren, Chris G.C. Jacobs, Elizabeth J. Duncan, David Armisén, Barbara M.I. Vreede, Patrice Baa-Puyoulet, Chloé S. Berger, Chun-che Chang, Hsu Chao, Mei-Ju M. Chen, Yen-Ta Chen, Christopher P. Childers, Ariel D. Chipman, Andrew G. Cridge, Antonin J.J. Crumière, Peter K. Dearden, Elise M. Didion, Huyen Dinh, harshavardhan doddapaneni, Amanda Dolan, Shannon Dugan-Perez, Cassandra G. Extavour, Gérard Febvay, Markus Friedrich, Neta Ginzburg, Yi Han, Peter Heger, Thorsten Horn, Yi-min Hsiao, Emily C. Jennings, J. Spencer Johnston, Tamsin E. Jones, Jeffery W. Jones, Abderrahman Khila, Stefan Koelzer, Viera Kovacova, Megan Leask, Sandra L. Lee, Chien-Yueh Lee, Mackenzie R. Lovegrove, Hsiao-Ling Lu, Yong Lu, Patricia J. Moore, Monica C. Munoz-Torres, Donna M. Muzny, Subba R. Palli, Nicolas Parisot, Leslie Pick, Megan Porter, Jiaxin Qu, Peter N. Refki, Rose Richter, Rolando Rivera-Pomar, Andrew J. Rosendale, Siegfried Roth, Lena Sachs, M. Emília Santos, Jan Seibert, Essia Sghaier, Jayendra N. Shukla, Richard J. Stancliffe, Olivia Tidswell, Lucila Traverso, Maurijn van der Zee, Séverine Viala, Kim C. Worley, Evgeny M. Zdobnov, Richard A. Gibbs, Stephen Richards
First draft genome assembly of an iconic clownfish species (Amphiprion frenatus). Anna Marcionetti, Victor Rossier, Joris A. M. Bertrand, Glenn Litsios, Nicolas Salamin
De novo assemblies of high-quality reference transcriptomes identifies Rosaceae-common and Rosa-specific encoding genes. Shubin Li, Micai Zhong, Xue Dong, Xiaodong Jiang, Yibo Sun, Dezhu Li, Kaixue Tang, Jin-Yong Hu
Analysis Of The Genomic Basis Of Functional Diversity In Dinoflagellates Using A Transcriptome-Based Sequence Similarity Network. Arnaud Meng, Erwan Corre, Ian Probert, Andres Gutierrez-Rodriguez, Raffaele Siano, Anita Annamale, Adriana Alberti, Corinne Da Silva, Patrick Wincker, Stephane Le Crom, Fabrice Not, Lucie Bittner
Molecular insights into the pathways underlying naked mole-rat eusociality. Eskeatnaf Mulugeta, Lucile Marion-Poll, David Gentien, Stefanie B. Ganswindt, Andre Ganswindt, Nigel C. Bennett, Elizabeth H. Blackburn, Chris G. Faulkes, Edith Heard
Long-lived rodents reveal signatures of positive selection in genes associated with lifespan and eusociality. Arne Sahm, Martin Bens, Karol Szafranski, Susanne Holtze, Marco Groth, Matthias Goerlach, Cornelis Calkhoven, Christine Mueller, Matthias Schwab, Hans Armin Kestler, Alessandro Cellerino, Hynek Burda, Thomas Hildebrandt, Philip Dammann, Matthias Platzer
Predicting the genetic loci of past evolution. Virginie Courtier-Orgogozo, Arnaud Martin
The Chromosomal Basis of Species Initiation: Prdm9 as an Anti-Speciation Gene. Donald R. Forsdyke
A most wanted list of conserved protein families with no known domains. Stacia K. Wyman, Aram Avila-Herrera, Stephen Nayfach, Katherine S. Pollard
Concomitant Duplication Of The Cid And Cenp-C Genes In Drosophila. Jose R. Teixeira, Guilherme B. Dias, Marta Svartman, Alfredo Ruiz, Gustavo C. S. Kuhn
Four new induced pluripotent stem cell lines produced from northern white rhinoceros with non-integrating reprogramming factors. Marisa L. Korody, Cullen Pivaroff, Thomas D. Nguyen, Suzanne E. Peterson, Oliver A. Ryder, Jeanne F. Loring
Heterochronic Developmental Shifts Underlie Floral Diversity within Jaltomata (Solanaceae). Jamie L. Kostyun, Jill C. Preston, Leonie C. Moyle
Reduction in major transcription factor binding domain families during the evolution of the avian lineage. Allie M. Graham, Jason S. Presnell
Egg viability decreases rapidly with time since ovulation in the rainbow darter: implications for the costs of choosiness. Rachel L. Moran, Rachel M. Soukup, Muchu Zhou, Rebecca C. Fuller
Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Samuel H. Lewis, Kaycee A. Quarles, Yujing Yang, Melanie Tanguy, Lise Frezal, Stephen A. Smith, Prashant P. Sharma, Richard Cordaux, Clement Gilbert, Isabelle Giraud, David H. Collins, Phillip D. Zamore, Eric A. Miska, Peter Sarkies, Francis M. Jiggins
Re-evaluating inheritance in genome evolution: widespread transfer of LINEs between species. Atma Ivancevic, Daniel Kortschak, Terry Bertozzi, David Adelson
Independent erosion of conserved transcription factor binding sites points to shared hindlimb, vision, and scrotum loss in different mammals. Mark J Berger, Aaron M Wenger, Harendra Guturu, Gill Bejerano
Cell biology
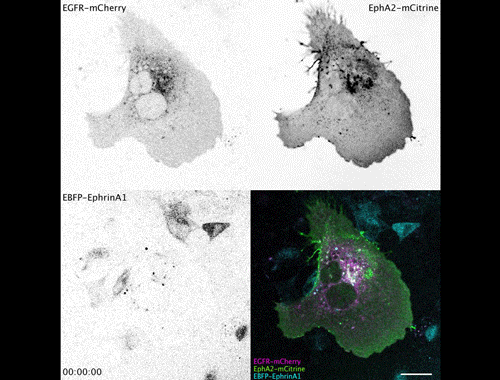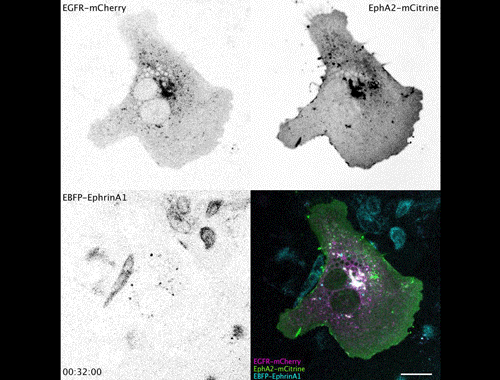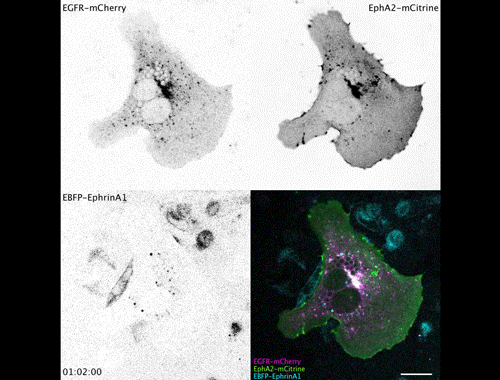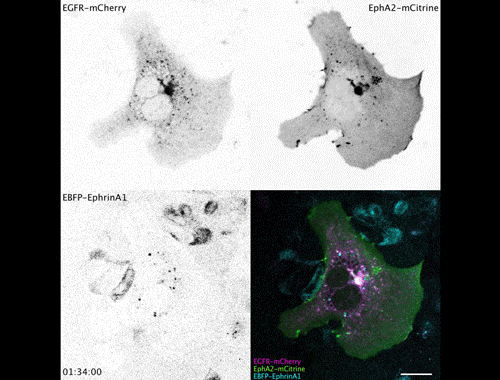
Contact inhibitory Eph signalling decouples EGFR activity from vesicular recycling to generate contextual plasticity. Wayne Stallaert, Ola Sabet, Yannick Bruggemann, Lisa Baak, Philippe Bastiaens
Characterization of Thylakoid Division Using Chloroplast Dividing Mutants in Arabidopsis. Jonathan Ho, Warren Kwan, Vivian Li, Steven Theg
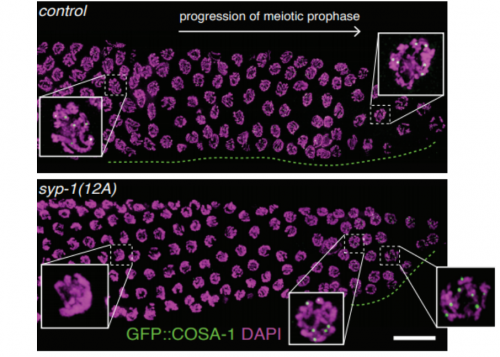
Phosphorylation of the synaptonemal complex protein SYP-1 promotes meiotic chromosome segregation. Aya Sato-Carlton, Chihiro Nakamura-Tabuchi, Stephane Kazuki Chartrand, Tomoki Uchino, Peter Mark Carlton
TPXL-1 Activates Aurora A to Clear Contractile Ring Components from the Polar Cortex During Cytokinesis. Sriyash Mangal, Jennifer Sacher, Taekyung Kim, Daniel Sampaio Osório, Fumio Motegi, Ana Xavier Carvalho, Karen Oegema, Esther Zanin
Myosin IIA interacts with the spectrin-actin membrane skeleton to control red blood cell membrane curvature and deformability. Alyson S Smith, Roberta B Nowak, Sitong Zhou, Michael Gianetto, David S Gokhin, Julien Papoin, Ionita C Ghiran, Lionel Blanc, Jiandi Wan, Velia M Fowler
Analysis of LRRC45 indicates cooperative functions of distal appendages at early steps of ciliogenesis. Bahtiyar Kurtulmus, Cheng Yuan, Jakob Schuy, Annett Neuner, Shoji Hata, Georgios Kalamakis, Ana Martin-Villalba, Gislene Pereira
Defining the Layers of a Sensory Cilium with STORM and Cryo-Electron Nanoscopies. Michael A. Robichaux, Valencia L. Potter, Zhixian Zhang, Feng He, Michael F. Schmid, Theodore G. Wensel
Mechanochemical feedback and control of endocytosis and membrane tension. Joseph Jose Thottacherry, Anita Joanna Kosmalska, Alberto Elosegui-Artola, Susav Pradhan, Sumit Sharma, Parvinder P. Singh, Marta C. Guadamillas, Natasha Chaudhary, Ram Vishwakarma, Xavier Trepat, Miguel A. del Pozo, Robert G. Parton, Pramod Pullarkat, Pere Roca-Cusachs, Satyajit Mayor
Morphological changes of plasma membrane and protein assembly during clathrin-mediated endocytosis. Aiko Yoshida, Nobuaki Sakai, Yoshitsugu Uekusa, Yuka Imaoka, Yoshitsuna Itagaki, Yuki Suzuki, Shige H. Yoshimura
Competition between microtubule-associated proteins directs motor transport. Brigette Y. Monroy, Danielle L. Sawyer, Bryce E. Ackermann, Melissa M. Borden, Tracy C. Tan, Kassandra M. Ori-McKenney
A size-invariant bud-length timer enables robustness in yeast cell size control. Corey A. H. Allard, Franziska Decker, Orion D. Weiner, Jared E. Toettcher, Brian R. Graziano
Cell-specific modulation of nuclear pore complexes controls cell cycle entry during asymmetric division. Arun Kumar, Priyanka Sharma, Zhanna Shcheprova, Anne Daulny, Trinidad Sanmartin, Irene Matucci, Charlotta Funaya, Miguel Beato, Manuel Mendoza
Local and global Cdc42 GEFs for fission yeast cell polarity are coordinated by microtubules and the Tea1/Tea4/Pom1 axis. Ye Dee Tay, Marcin Leda, Andrew B. Goryachev, Kenneth E. Sawin
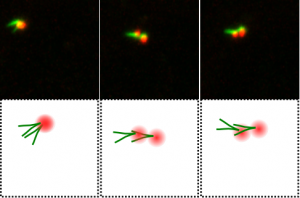
Stable centrosomal roots disentangle to allow interphase centriole independence. Robert Mahen
Temporal profiling of redox-dependent heterogeneity in single cells. Meytal Radzinski, Rosi Fassler, Ohad Yogev, William Breuer, Jenia Gutin, Tommer Ravid, Nir Friedman, Maya Schuldiner, Nadav Shai, Dana Reichmann
Modelling
Mathematical modelling of cortical neurogenesis reveals that the founder population does not necessarily scale with neuronal output. Noemi Picco, Fernando Garcia-Moreno, Thomas Woolley, Philip Maini, Zoltan Molnar
Single-Cell Reprogramming Of Mouse Embryo Development Through A Critical Transition State. Masa Tsuchiya, Alessandro Giuliani, Kenichi Yoshikawa
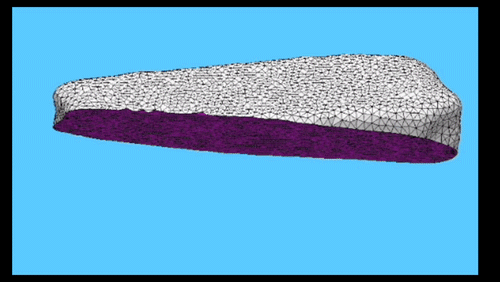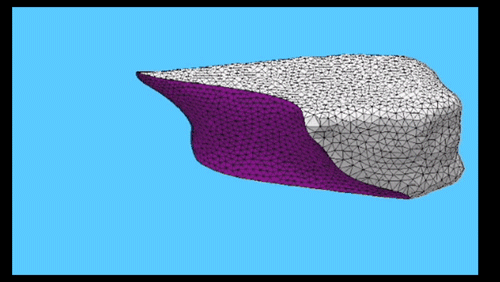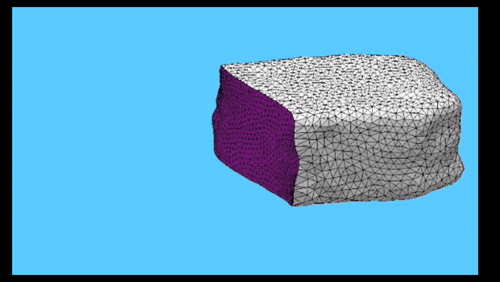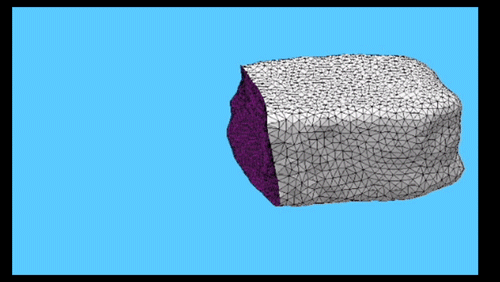
Using the three dimensional shape of plant cells to predict probabilities of cell division orientation. Pablo Martinez, Lindy Allsman, Kenneth A Brakke, Christopher Hoyt, Jordan Hayes, Carolyn G Rasmussen
Spot dynamics in a reaction-diffusion model of plant root hair initiation. Daniele Avitabile, Victor F. Brena-Medina, Michael J. Ward
Simulating Organogenesis in COMSOL: Tissue Mechanics. M. D. Peters, D. Iber
Simulating Organogenesis in COMSOL: Comparison Of Methods For Simulating Branching Morphogenesis. Lucas D. Wittwer, Michael Peters, Sebastian Aland, Dagmar Iber
A Dynamically Diluted Alignment Model Reveals the Impact of Cell Turnover on the Plasticity of Tissue Polarity Patterns. Karl B. Hoffmann, Anja Voss-Böhme, Jochen C. Rink, Lutz Brusch
Emerging whole-cell modeling principles and methods. Arthur P. Goldberg, Balázs Szigeti, Yin Hoon Chew, John A. P. Sekar, Yosef D. Roth, Jonathan R. Karr
A blueprint for human whole-cell modeling. Balazs Szigeti, Yosef D. Roth, John A. P. Sekar, Arthur P. Goldberg, Saahith C. Pochiraju, Jonathan R. Karr
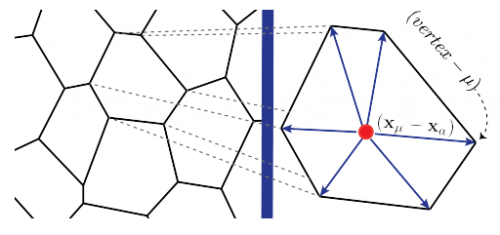
Hydrodynamics of shape-driven rigidity transitions in motile tissues. Michael Czajkowski, Dapeng Bi, M. Lisa Manning, M. Cristina Marchetti
Getting in shape and swimming: the role of cortical forces and membrane heterogeneity in eukaryotic cells. Hao Wu, Marco Avila Ponce de Leon, Hans Othmer
Inferring parameters for a lattice-free model of cell migration and proliferation using experimental data. Alexander Browning, Scott W. McCue, Rachelle N. Binny, Michael J. Plank, Esha T. Shah, Matthew J. Simpson
The self-organization of plant microtubules in three dimensions enables stable cortical localization and sensitivity to external cues. Vincent Mirabet, Pawel Krupinski, Olivier Hamant, Elliot Meyerowitz, Henrik Jonsson, Arezki Boudaoud
Universal Scaling in Biochemical Networks. Hyunju Kim, Harrison Smith, Cole Mathis, Jason Raymond, Sara Walker
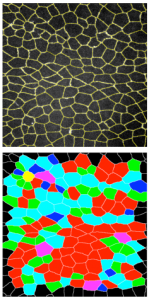
Universal statistics of epithelial tissue topology. Abdulaziz Abdullah, Demetris Avraam, Oleksandr Chepizhko, Thomas Vaccari, Stefano Zapperi, Caterina A. M. La Porta, Bakhtier Vasiev
A stochastic and dynamical view of pluripotency in mouse embryonic stem cells. Yen Ting Lin, Peter G. Hufton, Esther J. Lee, Davit A. Potoyan
Force-driven growth of intercellular junctions. Mohammad Tehrani, Alireza Sarvestani
Tools & resources
| Imaging etc.
A toolbox of anti-mouse and rabbit IgG secondary nanobodies. Tino Pleiner, Mark Bates, Dirk Goerlich
Dynamic and Selective Low-Complexity Domain Interactions Revealed by Live-Cell Single-Molecule Imaging. Shasha Chong, Claire Dugast-Darzacq, Zhe Liu, Peng Dong, Gina Dailey, Sambashiva Banala, Luke Lavis, Xavier Darzacq, Robert Tjian
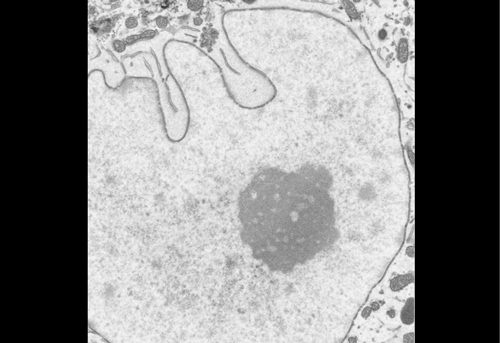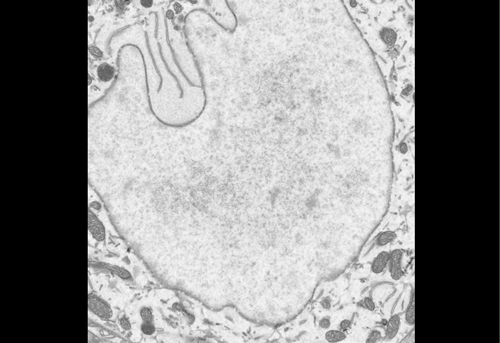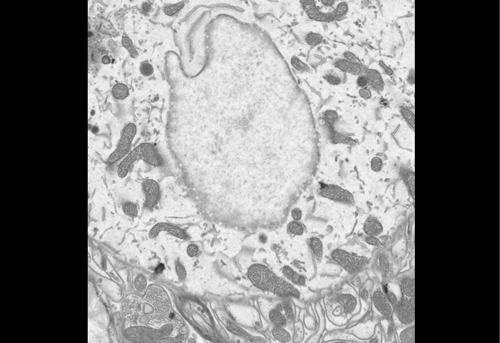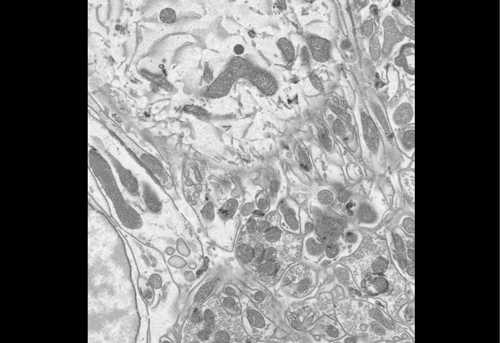
High contrast staining for serial block face scanning electron microscopy without uranyl acetate. Adolfo Odriozola, Jaime Llodrá, Julika Radecke, Céline Ruegsegger, Stefan Tschanz, Smita Saxena, Stephan Rohr, Benoît Zuber
Directed evolution of TurboID for efficient proximity labeling in living cells and organisms. Tess C Branon, Justin A Bosch, Ariana D Sanchez, Namrata D Udeshi, Tanya Svinkina, Steven A Carr, Jessica L Feldman, Norbert Perrimon, Alice Y Ting
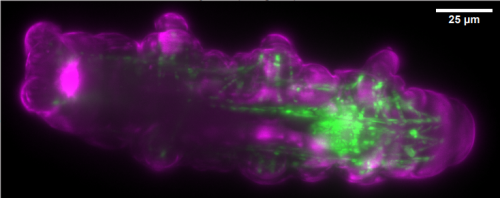
LITE microscopy: a technique for high numerical aperture, low photobleaching fluorescence imaging. Tanner C Fadero, Therese M Gerbich, Kishan Rana, Aussie Suzuki, Matthew DiSalvo, Kristina N Schaefer, Jennifer K Heppert, Thomas C Boothby, Bob Goldstein, Mark Peifer, Nancy L Allbritton, Amy S Gladfelter, Amy S Maddox, Paul S Maddox
Fluorescence polarization control for on-off switching of single molecules at cryogenic temperatures. Christiaan Hulleman, Maximiliaan Huisman, Robert Moerland, David Grunwald, Sjoerd Stallinga, Bernd Rieger
Image-based methods for phenotyping growth dynamics and fitness in large plant populations. François Vasseur, George Wang, Justine Bresson, Rebecca Schwab, Detlef Weigel
Simultaneous cell traction and growth measurements using light. Shamira Sridharan, Yanfen Li, Louis Foucard, Hassaan Majeed, Basanta Bhaduri, Alex Levine, Kristopher Kilian, Gabriel Popescu
Neocortical microdissection at columnar and laminar resolution for molecular interrogation. Koen Kole, Tansu Celikel
Two-photon photoactivated voltage imaging in tissue with an Archaerhodopsin-derived reporter. Miao-Ping Chien, Daan Brinks, Yoav Adam, William Bloxham, Simon Kheifets, Adam E. Cohen
Arabidopsis phenotyping through Geometric Morphometrics. Carlos A. Manacorda, Sebastian Asurmendi
3D cell nuclear morphology: microscopy imaging dataset and voxel-based morphometry classification results. Alexandr A. Kalinin, Ari Allyn-Feuer, Alex Ade, Gordon-Victor Fon, Walter Meixner, David Dilworth, Jeffrey R. de Wet, Gerald A. Higgins, Gen Zheng, Amy Creekmore, John W. Wiley, James E. Verdone, Robert W. Veltri, Kenneth J. Pienta, Donald S. Coffey, Brian D. Athey, Ivo D. Dinov
A spontaneously blinking fluorescent protein for simple single laser super-resolution live cell imaging. Yoshiyuki Arai, Hiroki Takauchi, Yuhei Ogami, Satsuki Fujiwara, Masahiro Nakano, Takeharu Nagai
48-spot single-molecule FRET setup with periodic acceptor excitation. Antonino Ingargiola, Maya Segal, Angelo Gulinatti, Ivan Rech, Ivan Labanca, Piera Maccagnani, Massimo Ghioni, Shimon Weiss, Xavier Michalet
Optical crosstalk in SPAD arrays for high-throughput single-molecule fluorescence spectroscopy. Antonino Ingargiola, Maya Segal, Angelo Gulinatti, Ivan Rech, Ivan Labanca, Piera Maccagnani, Massimo Ghioni, Shimon Weiss, Xavier Michalet
WorMachine: Machine Learning-Based Phenotypic Analysis Tool for Worms. Adam Hakim, Yael Mor, Itai Antoine Toker, Amir Levine, Yishai Markovitz, Oded Rechavi
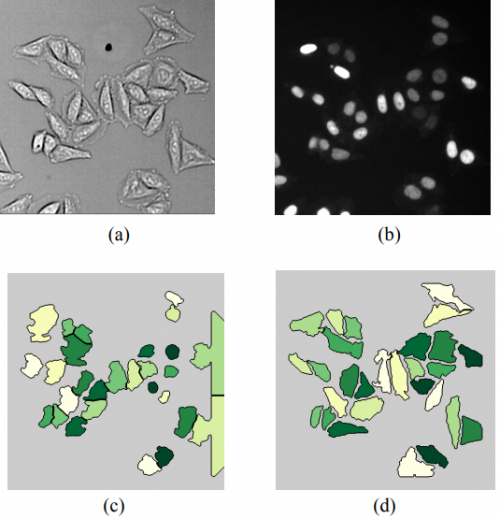
Active deep learning reduces annotation burden in automatic cell segmentation. Aritra Chowdhury, Simone Bianco, Sujoy Biswas
Magneto-active substrates for local mechanical stimulation of living cells. Cécile M Bidan, Mario Fratzl, Alexis Coullomb, Philippe Moreau, Alain H Lombard, Irène Wang, Martial Balland, Thomas Boudou, Nora M Dempsey, Thibaut Devillers, Aurélie Dupont
Light-Activated Cell Identification and Sorting (LACIS): A New Method to Identify and Select Edited Clones on a Microfluidic Device. Annamaria Mocciaro, Theodore Roth, Hayley Bennett, Magali Soumillon, Abhik Shah, Joseph Hiatt, Kevin Chapman, Alexander Marson, Gregory Lavieu
MS Western, a method of multiplexed absolute protein quantification is a practical alternative to western blotting. Mukesh Kumar, Shai R. Joseph, Martina Augsburg, Aliona Bogdanova, David Drechsel, Nadine L. Vastenhouw, Frank Buchholz, Marc Gentzel, Andrej Shevchenko
Obtaining 3D Super-resolution Information from 2D Super-resolution Images through a 2D-to-3D Transformation Algorithm. Andrew Ruba, Joseph Kelich, Wangxi Luo, Weidong Yang
Porcupine: a visual pipeline tool for neuroimaging analysis. Tim van Mourik,Lukas Snoek, Tomas Knapen, David Norris
Development of a riboswitch-based platform for live cell imaging of RNAs in mammalian cells. Esther Braselmann, Aleksandra J Wierzba, Jacob T Polaski, Mikołaj Chromiński, Dilara Batan, Dorota Gryko, Robert T Batey, Amy E Palmer
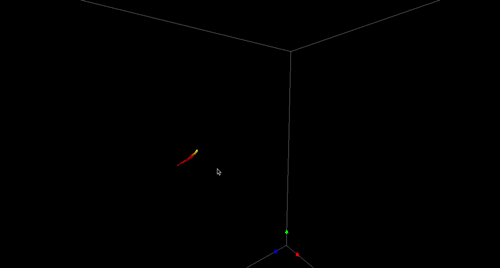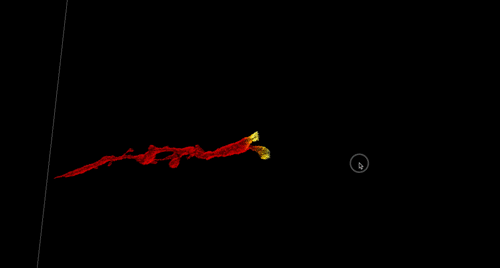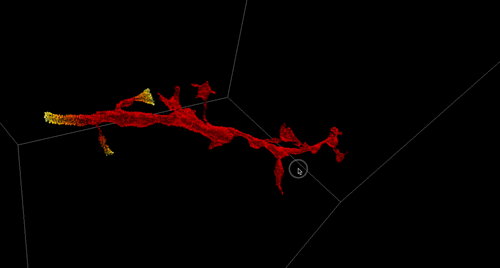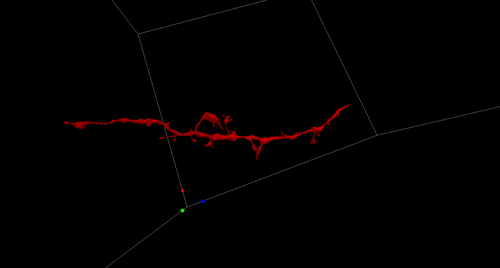
High-Precision Automated Reconstruction of Neurons with Flood-filling Networks. Michał Januszewski, Jörgen Kornfeld, Peter H Li, Art Pope, Tim Blakely, Larry Lindsey, Jeremy B Maitin-Shepard, Mike Tyka, Winfried Denk, Viren Jain
| Genome tools
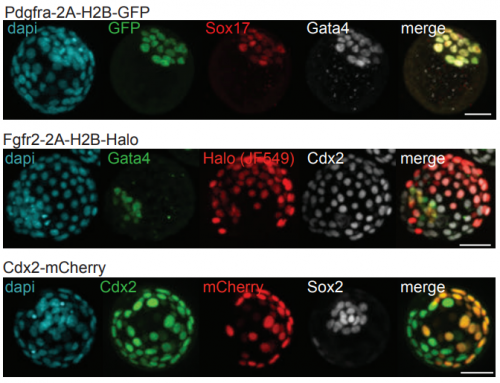
Efficient generation of targeted large insertions in mouse embryos using 2C-HR-CRISPR. Bin Gu, Eszter Posfai, Janet Rossant
Successful optimization of CRISPR/Cas9-mediated defined point mutation knock-in using allele-specific PCR assays in zebrafish. Sergey V. Prykhozhij, Charlotte Fuller, Shelby L. Steele, Chansey J. Veinotte, Babak Razaghi, Johane Robitaille, Christopher McMaster, Adam Shlien, David Malkin, Jason N. Berman
Efficient genome editing in primary cells and in vivo using viral-derived “Nanoblades” loaded with Cas9/sgRNA ribonucleoproteins. Philippe E. Mangeot, Valerie Rissons, Floriane Fusil, Aline Marnef, Emilie Laurent, Juliana Blin, Virginie Mournetas, Emmanuelle Massourides, Thibault J. M. Sohier, Antoine Corbin, Fabien Aube, Christian Pinset, Laurent Schaeffer, Gaelle Legube, Francois-Loic Cosset, Els Verhoeyen, Theophile Ohlmann, Emiliano P. Ricci
Rapid high-resolution measurement of DNA replication timing by droplet digital PCR. Dzmitry G. Batrakou, Emma D. Heron, Conrad A. Nieduszynski
The VT GAL4, LexA, and split-GAL4 driver line collections for targeted expression in the Drosophila nervous system. Laszlo Tirian, Barry Dickson
Genetic reagents for making split-GAL4 lines in Drosophila. Heather Dionne, Karen Hibbard, Amanda Cavallaro, Jui-Chun Kao, Gerald M Rubin

Droplet-based microfluidic analysis and screening of single plant cells. Ziyi Yu, Christian R. Boehm, Julian M. Hibberd, Chris Abell, Jim Haseloff, Steven J. Burgess, Ivan Reyna-Llorens
Programmable RNA recognition using a CRISPR-associated Argonaute. Audrone Lapinaite, Jennifer A. Doudna, Jamie Cate
CRISPR-DS: An efficient, low DNA input method for ultra-accurate sequencing. Daniela Nachmanson, Shenyi Lian, Elizabeth K. Schmidt, Michael J. Hipp, Kathryn T. Baker, Yuezheng Zhang, Maria Tretiakova, Kaitlyn Loubet-Senear, Brendan F. Kohrn, Jesse J. Salk, Scott R. Kennedy, Rosa Ana Risques
A critical comparison of technologies for a plant genome sequencing project. Pirita Paajanen, George Kettleborough, Elena Lopez-Girona, Michael Giolai, Darren Heavens, David Baker, Ashleigh Lister, Gail Wilde, Ingo Hein, Iain Macaulay, Glenn J. Bryan, Matthew D. Clark
iREAD: A Tool For Intron Retention Detection From RNA-seq Data. Hong-Dong Li, Cory C. Funk, Nathan D. Price
Amplification-free, CRISPR-Cas9 Targeted Enrichment and SMRT Sequencing of Repeat-Expansion Disease Causative Genomic Regions. Yu-Chih Tsai, David Greenberg, James Powell, Ida Hoijer, Adam Ameur, Maya Strahl, Ethan Ellis, Inger Jonasson, Ricardo Mouro Pinto, Vanessa Wheeler, Melissa L. Smith, Ulf Gyllensten, Robert Sebra, Jonas Korlach, Tyson A. Clark
An open library of human kinase domain constructs for automated bacterial expression. Steven K. Albanese, Daniel L. Parton, Sonya M. Hanson, Lucelenie Rodríguez-Laureano, Mehtap Işık, Julie M. Behr, Scott Gradia, Chris Jeans, Nicholas M. Levinson, Markus A. Seeliger, John D. Chodera
Efficient in situ barcode sequencing using padlock probe-based BaristaSeq. Xiaoyin Chen, Yu-Chi Sun, George M. Church, Je Hyuk Lee, Anthony M. Zador
Darwin Assembly: fast, efficient, multi-site bespoke mutagenesis. Christopher Cozens, Vitor B. Bernardes Pinheiro
Gene-expression profiling of single cells from archival tissue with laser-capture microdissection and Smart-3SEQ. Joseph W Foley, Chunfang Zhu, Philippe Jolivet, Shirley X Zhu, Peipei Lu, Michael J Meaney, Robert B West
A duplex MIPs-based biological-computational cell lineage discovery platform. Liming Tao, Ofir Raz, Zipora Marx, Tamir Biezuner, Shiran Amir, Lilach Milo, Rivka Adar, Amos Onn, Noa Chapal-Ilani, Veronika Berman, Ron Levy, Barak Oron, Ehud Shapiro
Genome-Wide Mining, Characterization and Development of miRNA-SSRs in Arabidopsis thaliana. Anuj Kumar, Aditi Chauhan, Sai Kumar Kompelli, Vijay Gahlaut, Johny Ijaq, Krishna Pal Singh, MNV Prasad Gajula, Prashanth Suravajhala, Amrendra Mishra, Harindra Singh Balyan, Pushpendra Kumar Gupta
Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database. Luke Zappia, Belinda Phipson, Alicia Oshlack
BoostMe accurately predicts DNA methylation values in whole-genome bisulfite sequencing of multiple human tissues. Luli S. Zou, Michael R. Erdos, D. Leland Taylor, Peter S. Chines, Arushi Varshney, Stephen C. J. Parker, Francis S. Collins, John P. Didion

Rapid Agrobacterium-mediated transformation of tobacco cotyledons using toothpicks. Yuan-Yeu Yau, Mona Easterling, Lindsey Brennan
Robust identification of deletions in exome and genome sequence data based on clustering of Mendelian errors. Kathryn B. Manheimer, Nihir Patel, Felix Richter, Joshua Gorham, Angela C. Tai, Jason Homsy, Marko T. Boskovski, Michael Parfenov, Elizabeth Goldmuntz, Wendy K. Chung, Martina Brueckner, Martin Tristani-Firouzi, Deepak Srivastava, Jonathan G. Seidman, Christine E. Seidman, Bruce D. Gelb, Andrew J. Sharp
Mammalian genomic regulatory regions predicted by utilizing human genomics, transcriptomics and epigenetics data. Quan H. Nguyen, Ross L. Tellam, Marina Naval-Sanchez, Laercio R Porto-Neto, William Barendse, Antonio Reverter, Benjamin J Hayes, James W Kijas, Brian P. Dalrymple
Summarizing Performance for Genome Scale Measurement of miRNA: Reference Samples and Metrics. P. Scott Pine, Steven P. Lund, Jerod R. Parsons, Lindsay K. Vang, Ashish A. Mahabal, Luca Cinquini, Sean C. Kelly, Heather Kincaid, Daniel J. Crichton, Avrum Spira, Gang Liu, Adam C. Gower, Harvey I. Pass, Chandra Goparaju, Steven M. Dubinett, Kostyantyn Krysan, Sanford A. Stass, Debra Kukuruga, Kendall Van Keuren-Jensen, Amanda Courtright-Lim, Karol L. Thompson, Barry A. Rosenzweig, Lynn Sorbara, Sudhir Srivastava, Marc L. Salit
Predicting microRNA targeting efficacy in Drosophila. Vikram Agarwal, Alexander O. Subtelny, Prathapan Thiru, Igor Ulitsky, David P. Bartel
Immuno-Detection by sequencing (ID-seq) enables large-scale high-dimensional phenotyping in cells. Jessie A. van Buggenum, Jan P. Gerlach, Sabine E. J. Tanis, Mark Hogeweg, Jesse Middelwijk, Ruud van der Steen, Cornelis A. Albers, Klaas W. Mulder
In vivo RNA targeting of point mutations via suppressor tRNAs and adenosine deaminases. Dhruva Katrekar, Prashant Mali
High-frequency off-target mutagenesis induced by CRISPR/Cas9 in Arabidopsis and its prevention by improving specificity of the tools. Qiang Zhang, Hui-Li Xing, Zhi-Ping Wang, Hai-Yan Zhang, Fang Yang, Yun Zhou, Xue-Chen Wang, Qi-Jun Chen
Single Molecule Sequencing of Cell-free DNA from Maternal Plasma for Noninvasive Trisomy Detection. Minyue Dong, Liwei Deng, Huan Jin, Jinsen Cai, Huan Shang, Shuo Zhang, Yueping Zhang, Jinzhou Qin, Dikai Zhang, Feng Long, Renli Zhang, Runsheng Chen, Michael W. Deem, Jun Yu, Jiankui He
Graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. F. Alexander Wolf, Fiona Hamey, Mireya Plass, Jordi Solana, Joakim S. Dahlin, Berthold Gottgens, Nikolaus Rajewsky, Lukas Simon, Fabian J. Theis
Linked-read analysis identifies mutations in single-cell DNA sequencing data. Craig L Bohrson, Alison R Barton, Michael A Lodato, Rachel E Rodin, Vinay Viswanadham, Doga Gulhan, Isidro Cortes, Maxwell A Sherman, Lovelace J Luquette, Minseok Kwon, Michael E Coulter, Christopher A Walsh, Peter J Park
DiscoSnp++: de novo detection of small variants from raw unassembled read set(s). Pierre Peterlongo, Chloe Riou, Erwan Drezen, Claire Lemaitre
A Unified Statistical Framework for Single Cell and Bulk RNA Sequencing Data. Lingxue Zhu, Jing Lei, Bernie Devlin, Kathryn Roeder
ACEseq – allele specific copy number estimation from whole genome sequencing. Kortine Kleinheinz, Isabell Bludau, Daniel Huebschmann, Michael Heinold, Philip Kensche, Zuguang Gu, Cristina Lopez, Michael Hummel, Wolfram Klapper, Peter Moeller, Inga Vater, Rabea Wagener, ICGC MMML-Seq project, Benedikt Brors, Reiner Siebert, Roland Eils, Matthias Schlesner
Adapterama III: Quadruple-indexed, triple-enzyme RADseq libraries for about $1USD per Sample (3RAD). Travis C. Glenn, Natalia J. Bayona-Vasquez, Troy J. Kieran, Todd W. Pierson, Sandra L. Hoffberg, Peter A. Scott, Kerin E. Bentley, John W. Finger Jr., Patrick R. Watson, Swarnali Louha, Nick Troendle, Pindaro Diaz-Jaimes, Rodney Mauricio, Brant C. Faircloth
HiGlass: Web-based Visual Exploration and Analysis of Genome Interaction Maps. Peter Kerpedjiev, Nezar Abdennur, Fritz Lekschas, Chuck McCallum, Kasper Dinkla, Hendrik Strobelt, Jacob M. Luber, Scott B. Ouellette, Alaleh Azhir, Nikhil Kumar, Jeewon Hwang, Soohyun Lee, Burak H. Alver, Hanspeter Pfister, Leonid A. Mirny, Peter J. Park, Nils Gehlenborg
miCloud: a plug and play, on-premises bioinformatics cloud, providing seamless integration with Illumina genome sequencers. Baekdoo Kim, Thamina Ali, Konstantinos Krampis, Changsu Dong, Bobby Laungani, Claudia Wultsch, Carlos Lijeron
Data quality of Whole Genome Bisulfite Sequencing on Illumina platforms. Amanda Raine, Ulrika Liljedahl, Jessica Nordlund
Systematic evaluation of statistical methods for identifying looping interactions in 5C data. Jennifer E Phillips-Cremins, Thomas G Gilgenast
A novel feature selection for RNA-seq analysis. Henry Han
Identification of Cell States Using Super-Enhancer RNA. Yueh-Hua Tu, Hsueh-Fen Juan, Hsuan-Cheng Huang
HebbPlot: An intelligent tool for learning and visualizing chromatin mark signatures. Hani Z Girgis, Alfredo Velasco II
INFERNO – INFERring the molecular mechanisms of NOncoding genetic variants. Alexandre Amlie-Wolf, Mitchell Tang, Elisabeth E. Mlynarski, Pavel P. Kuksa, Otto Valladares, Zivadin Katanic, Debby Tsuang, Christopher D. Brown, Gerard D. Schellenberg, Li-San Wang
Revising transcriptome assemblies with phylogenetic information in Agalma1.0. August Guang, Mark Howison, Felipe Zapata, Charles E. Lawrence, Casey Dunn
The Bio::Phylo libraries for phylogenetic data analysis, version 2.0. Rutger A. Vos, Hannes Hettling
SALP, a new single-stranded DNA library preparation method especially useful for the high-throughput characterization of chromatin openness states. Jian Wu, Wei Dai, Ling Wu, Jinke Wang
RACIPE: A computational tool for Modeling Gene Regulatory Circuits using Randomization. Bin Huang, Dongya Jia, Jingchen Feng, Herbert Levine, Jose N. Onuchic, Mingyang Lu
VASC: dimension reduction and visualization of single cell RNA sequencing data by deep variational autoencoder. Dongfang Wang, Jin Gu
C3D: A tool to predict 3D genomic interactions between cis-regulatory elements. Tahmid Mehdi, Swneke D Bailey, Paul Guilhamon, Mathieu Lupien
QTLseqr: An R package for bulk segregant analysis with next-generation sequencing. Ben Mansfeld, Rebecca Grumet
Modeling transcription factor combinatorics in promoters and enhancers. Jimmy Vandel, Oceane Cassan, Sophie Lebre, Charles-Henri Lecellier, Laurent Brehelin
Research practice
Bioconda: A sustainable and comprehensive software distribution for the life sciences. Björn Grüning, Ryan Dale, Andreas Sjödin, Jillian Rowe, Brad A. Chapman, Christopher H. Tomkins-Tinch, Renan Valieris, The Bioconda Team, Johannes Köster
Barriers to Integration of Bioinformatics into Undergraduate Life Sciences Education. Jason Williams, Jennifer Drew, Sebastian Galindo-Gonzalez, Srebrenka Robic, Elizabeth Dinsdale, William Morgan, Eric Triplett, James Burnette, Sam Donovan, Sarah Elgin, Edison Fowlks, Anya Goodman, Neal Grandgenett, Carlos Goller, Charles Hauser, John R. Jungck, Jeffrey Newman, William Pearson, Elizabeth Ryder, Melissa Wilson Sayres, Michael Sierk, Todd Smith, Rafael Tosado-Acevedo, William Tapprich, Tammy Tobin, Arlin Toro-Martínez, Lonnie Welch, Robin Wright, David Ebenbach, Mindy McWilliams, Anne Rosenwald, Mark Pauley
Practical computational reproducibility in the life sciences. Björn Grüning, John Chilton, Johannes Köster, Ryan Dale, Jeremy Goecks, Rolf Backofen, Anton Nekrutenko, James Taylor
A Data Citation Roadmap for Scholarly Data Repositories. Martin Fenner, Mercè Crosas, Jeffrey Grethe, David Kennedy, Henning Hermjakob, Philippe Rocca-Serra, Gustavo Durand, Robin Berjon, Sebastian Karcher, Maryann Martone, Timothy Clark
The uncertainty of old aliquots of cell lines: e-CAS or m-CAS? Elizabeth Evans, Romain Paillot, Maria Rocio Lopez-Alvarez
Scientific Theories and Artificial Intelligence. Philippe Desjardins-Proulx, Timothée Poisot, Dominique Gravel
Sharing genetic admixture and diversity of public biomedical datasets.Olivier Harismendy, Jihoon Kim, Xiaojun Xu, Lucila Ohno-Machado
Becoming more transparent: Collecting and presenting data on biomedical Ph.D. alumni. Christopher L Pickett, Shirley Tilghman
Publiphilia Impactfactorius: a new psychiatric syndrome among biomedical scientists? Joeri K Tijdink, Yvo M Smulders, Lex M Bouter
Idea farming: it is a good idea to have bad ideas in science. Christopher J Lortie
Sci-Hub provides access to nearly all scholarly literature. Daniel S Himmelstein, Ariel R Romero, Stephen R McLaughlin, Bastian Greshake Tzovaras, Casey S Greene
The Reproducibility Of Research And The Misinterpretation Of P Values. David Colquhoun
Why not…
Fluid mediated self-organization of ventilation in honeybee nests. Jacob M. Peters, Orit Peleg, L. Mahadevan
The scent of the fly. Paul G. Becher, Sebastien Lebreton, Erika A. Wallin, Erik Hedenstrom, Felipe Borrero-Echeverry, Marie Bengtsson, Volker Jorger, Peter Witzgall


 (No Ratings Yet)
(No Ratings Yet)