Spider segmentation gets its SOX on!
Posted by chrisbonatto, on 15 October 2018
There is a vast amount of information known about how some animals pattern their bodies into repeated segments, especially in the fruit-fly Drosophila melanogaster. However, when compared to other arthropods, there are several characteristics that are derived in the fruit fly. It has a very short development time, a syncytium at the blastoderm stage allowing molecules free passage throughout the embryo and the segments are generated almost simultaneously. This is called the “long germ band” type of arthropod development.
While the fruit fly undergoes “long germ band” development, most arthropods belong to the “short germ band” type, in which the head and a small portion of the trunk are generated after gastrulation, but posterior segments are then added one by one from the so called segment addition zone (SAZ). An earlier article on the Node previously outlined the reasons the spider is an excellent study organism (https://thenode.biologists.com/day-life-spider-lab/lablife/) – but we want to highlight a recent discovery that builds to this earlier story: some chelicerates have undergone a whole genome duplication during their evolutionary history (Schwager et al., 2017; Leite et al., 2018). This genome duplication event has allowed some of the conserved sets of genes to retain duplicated copies, potentially having huge implications for the regulation and evolution of developmental processes in these animals – including segmentation.
What do we know about segmentation in the common house spider Parasteatoda tepidariorum? A remarkable discovery in this species was that the Delta-Notch and Wnt signaling pathways were involved in the differentiation of the cells of the SAZ. These findings shifted the view of canonical segmentation in arthropods, since these two pathways are not involved in long germ development of the fruit fly (for further information, see Oda et al., 2007 and Schonauer et al., 2016).
Strikingly, we found in 2017 that another conserved metazoan gene family has retained duplicated copies in Parasteatoda (Paese et al., 2017), the Sox genes (Sry-related HMG Box). In this study, we isolated 14 Sox genes, analysed their expression patterns at different stages of embryonic development (Paese et al., 2017) and studied their functional role with RNA interference (Paese et al., 2018). Interestingly, some of these genes have no detectable expression during embryonic development, and as noted in other studies, Sox genes have a high degree of redundancy (i.e. genes with conserved motifs that therefore can act in the place of the missing gene) and we only observed an obvious phenotype in one out of fourteen genes, a SoxB gene – Sox21b-1. (Expression pattern in Figure 1). This is an example of one of the genes that have undergone a duplication – in which each copy is dispersed in the genome of the spider.
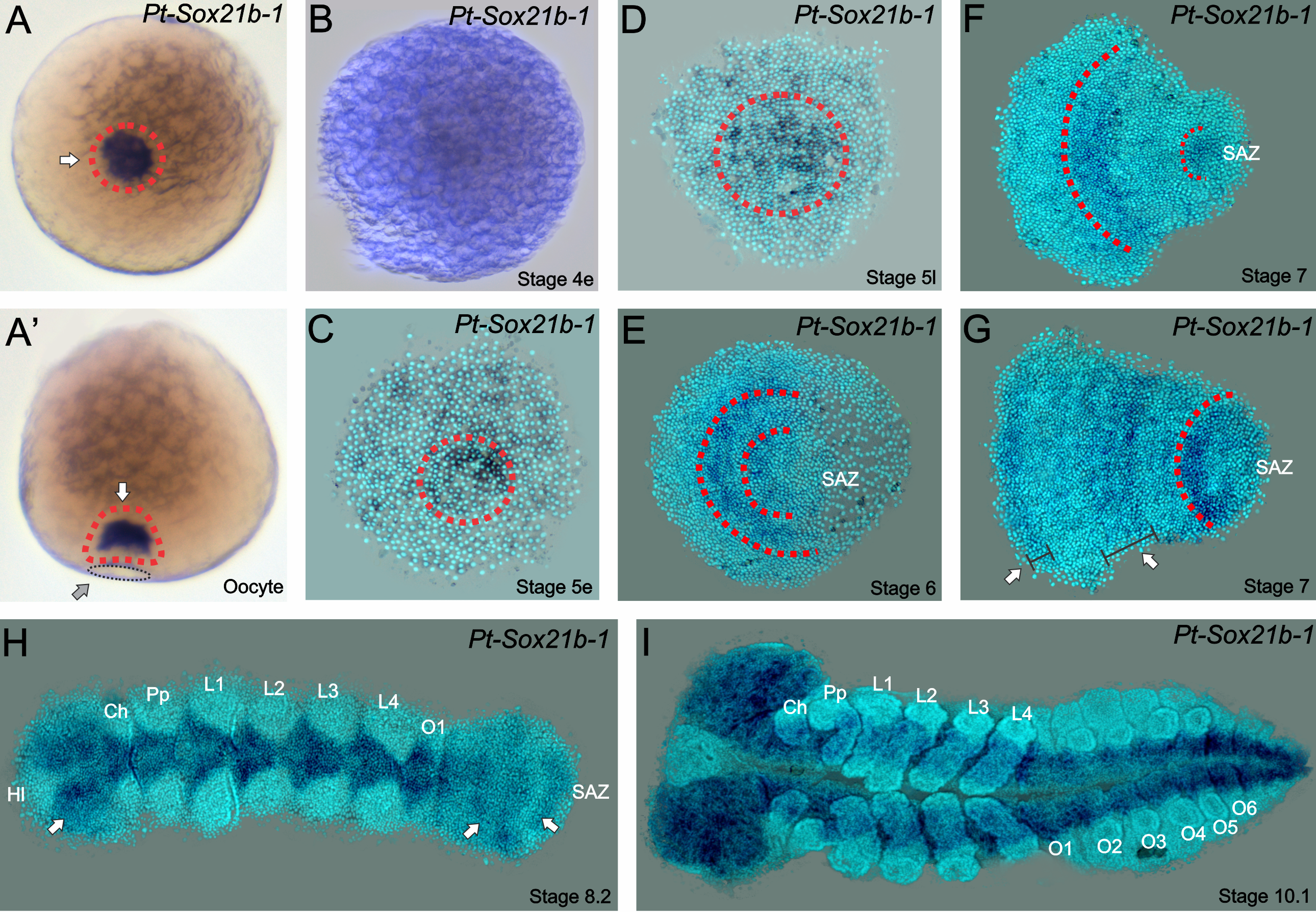
Figure 1
Even though the gene’s name may not be the most memorable when compared to some genes in the fruit-fly, the phenotype when Sox21b-1 is knocked down is very unforgettable. Moreover, this gene has an expression pattern similar to what is seen with a related gene, Dichaete, in other arthropods – which has no detectable expression during spider embryogenesis (for more info, see Paese et al., 2017). For a quick spoiler: we first thought that Sox21b-1 had acquired Dichate function in this spider species. Wouldn’t that be amazing? And how would we be sure of that? Let’s do some functional analysis!
When we injected double-stranded RNA in the opisthosoma of a female spider, we obtained phenotypes of different penetrance. During one rainy night in Oxford, I was analyzing a dish with embryos that I removed from a cocoon that was deposited by a Sox21-b1 dsRNA injected female. Using a stereomicroscope, I was only able to identify the head region and a very strange single pair of appendages growing from where six pairs should be growing. At a first glance, I was very excited about finding a new phenotype, but kept searching for other embryos with the same pattern in different cocoons. Sadly, in this first injection round, only two injected females survived – however, a significant proportion of embryos displayed a phenotype.
I injected another seven spiders and the phenotypes matched the first round of injections (Figure 2 for the phenotypes). In class I phenotypes, most embryos displayed only the first three pairs of appendages (namely chelicerae, pedipalps and leg bearing segment 1 – L1) instead of six pairs, whereas class II lack the L1 segment and class III did not develop further after gastrulation. Much more interesting than lacking all the leg-bearing segments was that this anterior region was able to differentiate even in the absence of a developing SAZ! These findings are further evidence for the hypothesis that the short germ arthropods mechanism for anterior and posterior segmentation are decoupled.
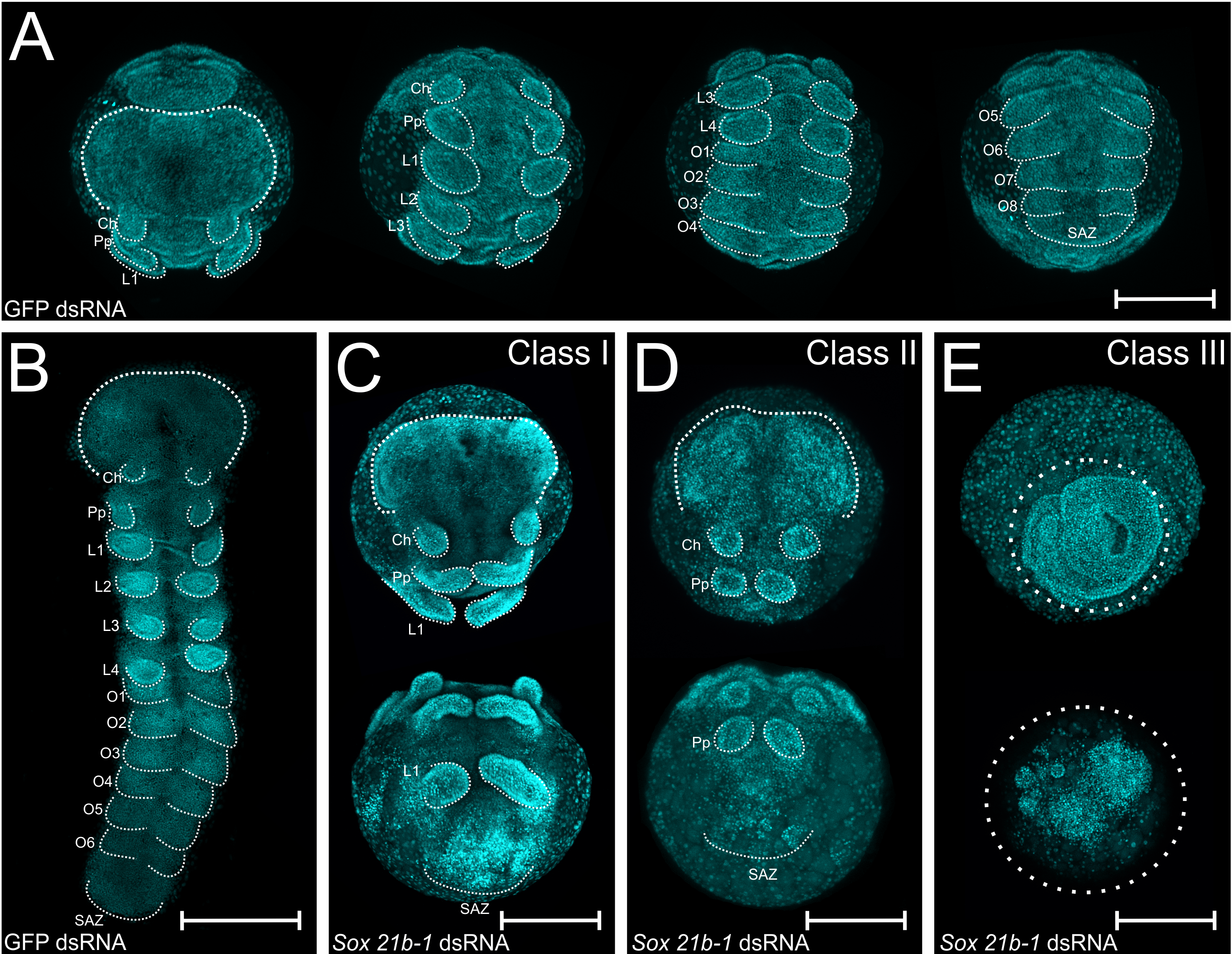
Figure 2
Knowing this, we started analyzing which mechanisms were affected by the knockdown of this Sox gene. First, we assayed the expression of known cumulus formation and migration genes: decapentaplegic and Ets4. The expression of both genes was normal (Figure 3 of the paper – A to D). Then the next targets to be evaluated were genes involved with secondary layer formation (forkhead) and head patterning (hedgehog). Whereas the expression of the latter was normal, forkhead expression was completely lost. This information led us to analyse determinants of mesoderm and endoderm formation in older embryos and we indeed observed an effect on those two layers in the Sox21b-1 phenotype. However, we cannot be sure if this is a direct effect due to the lack of Sox21b-1 or caused by the overall lower number of cells, which has to be investigated further.
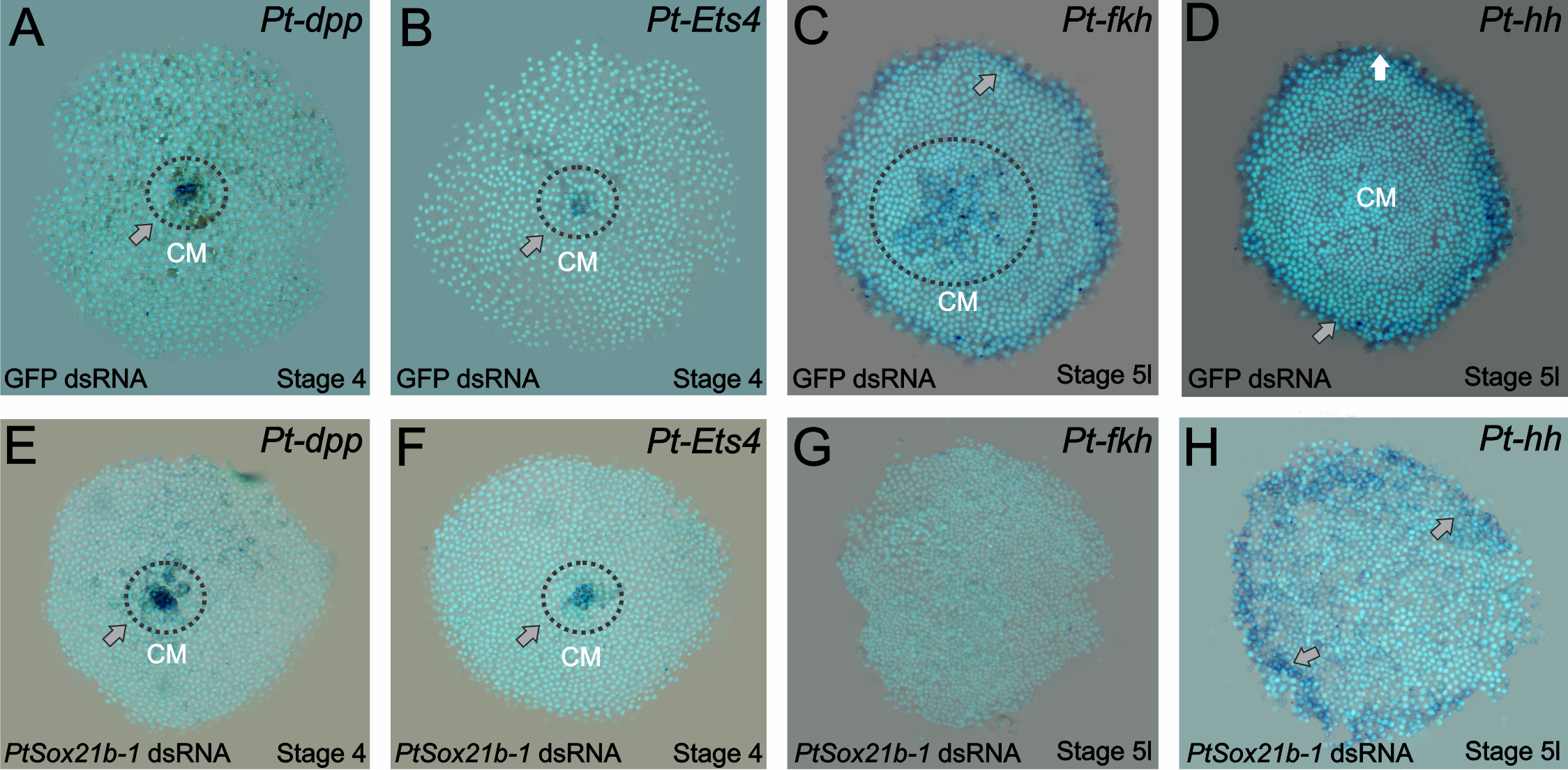
Figure 3
As mentioned before, the spider patterns its segments not following the Drosophila canonical mode, but instead through control by Delta-Notch and Wnt signaling, regulating pair-rule gene expression via caudal. Thus, we looked into the effect on expression of the known genes that are related to this mechanism in the spider. Interestingly, the expression of caudal, delta, hairy and wnt8 is lost in the SAZ, but we could still see the expression of the segment markers engrailed and hedgehog in the knockdown embryos. In summary, this means that this Sox gene is upstream of Wnt and Delta/Notch and this is additional evidence for the independence of anterior and posterior segmentation in the spider (Figure 4).
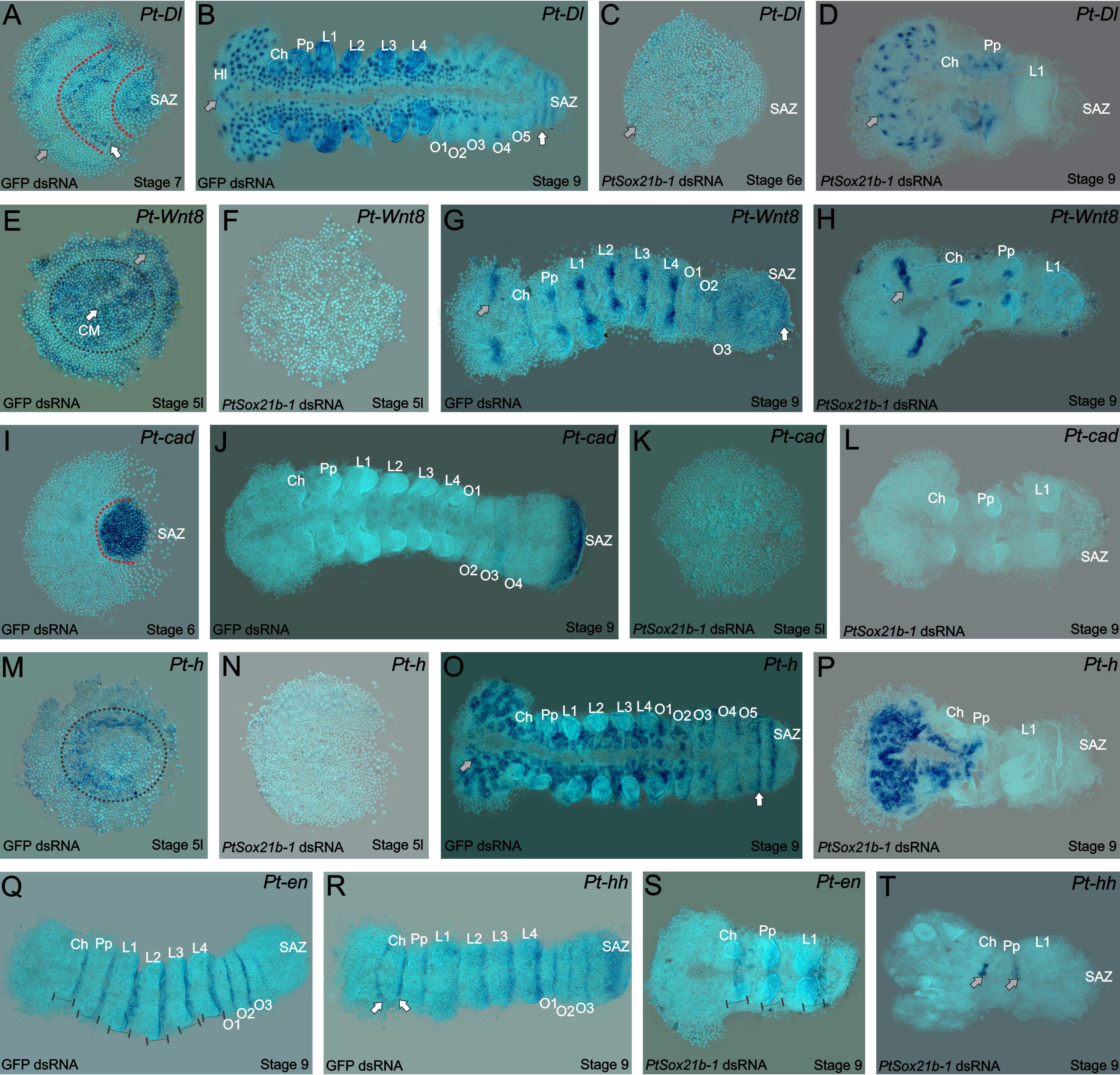
Figure 4
In conclusion, our study adds an important piece towards understanding segmentation in short germ arthropods and the effects of a whole genome duplication on developmental processes (Figure 5).
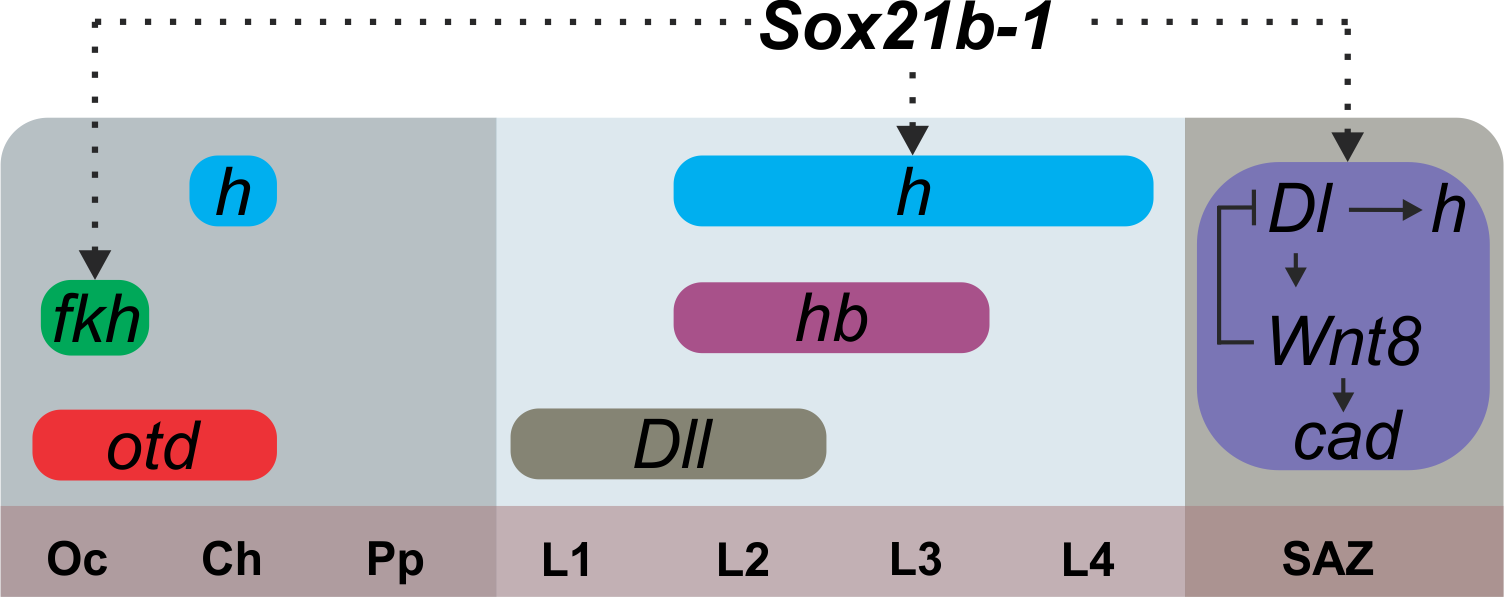
Figure 5
We might be far from a complete picture of how segmentation is regulated in spiders, but it is indeed exciting that there are many more pieces of the puzzle to be solved!
Supplementary References:
Leite, D. J., Baudouin-Gonzalez, L., Iwasaki-Yokozawa, S., Lozano-Fernandez, J., Turetzek, N., Akiyama-Oda, Y., Prpic, N. M., Pisani, D., Oda, H., Sharma, P. P., McGregor, A. P. (2018). Homeobox gene duplication and divergence in arachnids. Mol Biol Evol.
Oda H et al (2007) Progressive activation of Delta-Notch signaling from around the blastopore is required to set up a functional caudal lobe in the spider Achaearanea tepidariorum. Development, 134: 2195-2205.
Paese CLB, (2017) Duplication and divergence of Sox genes in spiders. bioRxiv.
Paese, CLB.; Schoenauer A, Leite DJ, Russell S., McGregor AP. (2018). A SoxB gene acts as an anterior gap gene and regulates posterior segment addition in a spider. eLife 2018;7:e37567
Schönauer A, et al. (2016) The Wnt and Delta-Notch signalling pathways interact to direct even-skipped expression via caudal during segment addition in the spider Parasteatoda tepidariorum. Development 143:2455-2463.
Schwager EE, et al. (2017) The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol 15(1):62.


 (3 votes)
(3 votes)