The people behind the papers – Pauline Anne & Christian Hardtke
Posted by the Node Interviews, on 25 May 2018
Short CLE peptides regulate a wide variety of processes during plant development. In the developing root, the receptors and co-receptors for CLEs have remained largely unclear, as have the relationships between different CLEs and different (co-)receptors. A new paper in Development addresses this problem by reporting a new receptor kinase involved in CLE signaling. We caught up with lead author Pauline Anne and her PI Christian Hardtke, Professor in the Department of Plant Molecular Biology at University of Lausanne, to find out more.

Christian, can you give us your scientific biography and the questions your lab is trying to answer?
Out of an early interest in genetics, I studied Biology at the University of Munich. I became particularly fascinated by Developmental Biology when I learned about the then still recent isolation of the Bicoid morphogen. At that point I was determined to work with Drosophila. Incidentally, Thomas Berleth, who had cloned Bicoid a few years earlier, worked with Gerd Juergens in the Genetics department at that time. They were recruiting an M.Sc. student to investigate the “Arabidopsis embryo patterning mutant monopteros”. I only realised during the interview that Arabidopsis was not some short germ-band insect, but an upcoming plant model organism. Maybe my judgement was clouded by the dense pipe smoke in Gerd’s office, but I committed to the project, which also became my Ph.D. In hindsight, it was a stroke of luck, I enjoy working with plants ever since.
Four years later I had cloned MONOPTEROS and walked enough along chromosomes. Thus, after briefly wrapping up my Ph.D. work at the University of Toronto with Thomas, I started my postdoc as an HFSP fellow in the lab of Xing-Wang Deng at Yale University, to learn biochemistry. It was an exciting time as the lab discovered the mechanistic basis of the COP9-COP1 signalosome-ubiquitin ligase system in Arabidopsis photomorphogenesis, and its operational conservation but differential output in distant eukaryotes. When I started my own lab at McGill University however, I turned to root system architecture with the aim to analyse its intraspecific natural genetic variation. Since then, we isolated several QTL alleles. Because I like to explore the molecular foundations of their developmental phenotypes, my lab’s focus has shifted in the years after I moved to the University of Lausanne. Nowadays we are mainly interested in deciphering molecular pathways that govern the patterned differentiation of plant vascular tissues, and in identifying the non-cell autonomous signals that are emitted from phloem vasculature to organize surrounding tissues.
And Pauline, how did you come to work in Christian’s lab and be involved with this project?
Christian and I share common interests. In my PhD, I characterised the phloem regulator OCTOPUS (OPS) and its interaction with the brassinosteroid phytohormone pathway. I wanted to continue working on phloem differentiation, and Christian is a leader in that field. We discussed possibilities, I wanted to continue exploring signalling pathways, so he proposed to analyse CLE26 sensing, which we suspected to play a role in phloem differentiation.
When did your lab first get interested in CLE peptide signalling in the root?
CH When we got stuck in our analysis of BREVIS RADIX (BRX), another positive regulator of phloem formation. Stephen Depuydt, a postdoc in my lab at the time, proposed to screen for suppressors of the phloem defects in brx mutants. Although I was rather sceptical about the prospect of healing the strong brx phenotype by second site mutation.
PA But it worked! Loss-of-function in BARELY ANY MERISTEM 3 (BAM3), a receptor kinase, was isolated as a perfect brx suppressor from this screen. They later found that BAM3 is the receptor for the phloem-specific CLE45 peptide, pointing to a role of autocrine CLE peptide signalling in phloem differentiation.
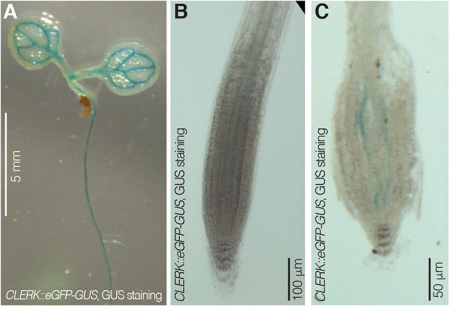
Can you give us the key results of the paper in a paragraph?
PA We isolated CLERK as a genetic component of CLE26 sensing in the root, but it is also required for perception of other CLE peptides. Moreover, CLERK encodes a co-receptor protein that acts in the phloem and is necessary to prevent premature protophloem specification.
CH Also, CLERK acts genetically in parallel to described CLE sensing pathways in the protophloem, corroborating the redundancy we suspected at the outset.
You have lots of CLE peptides, lots of receptors, lots of co-receptors, and lots of promiscuous receptor-ligand and receptor-co-receptor interactions. How does this system achieve any degree of specificity?
PA Although many exogenously applied CLE peptides suppress root protophloem differentiation, only few, like CLE45 or CLE26, are actually expressed in root development. Recent studies by colleagues suggest that some CLEs are induced by specific environmental stimuli, to shut down root growth in unfavourable conditions. So part of the answer probably lies in specific ligand expression.
CH …and in specific receptor and co-receptor expression. It is also noteworthy that some receptors are not only more promiscuous with respect to ligands than others, but that ligand binding affinities can vary by an order of magnitude. So even in presence of multiple receptor-coreceptor pairs, ligand interaction can likely trigger vastly different signal outputs. It will be a major challenge to map these interactions in time and space, also because cross-regulation between CLE-sensing receptors has been observed.
How might CLERK help you in future analysis of the CLE sensing pathway?
CH One surprising discovery was that in brx or ops mutants, CLERK is not required for CLE26 or CLE45 sensing. We can therefore perform a sensitized screen for CLE resistance, for example in a brx clerk double mutant background. This should enable us to home in on the CLE26 receptor, as well as receptors for other CLEs.
PA The system is more and more complex, and genetics does not always give the expected answers. But it looks like it is less complex in the root than in the shoot. For example, while our paper was under review, colleagues found that CLERK also plays a role in the shoot. However, there its action is masked by redundancy between four related co-receptors.
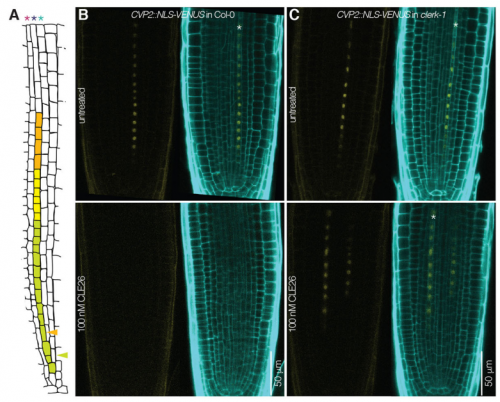
When doing the research, did you have any particular result or eureka moment that has stuck with you?
PA Yes, the day I received the whole genome sequencing results of our original clerk mutant! I was on vacation in Italy. As you guess, I could not wait to be back in the lab to analyse the results…and At2g23950 popped up, a receptor-like kinase. This was the best candidate I could expect at that moment.
And what about the flipside: any moments of frustration or despair?
PA Of course! Even if the project went well, there are more instances of frustration in research life than eureka moments. I think my main frustration was the very weak expression level of CLERK in the root tip. But on the upside, this forced me to hone my confocal microscopy skills…
What next for you after this paper?
PA In the short term, I will focus on the characterization of other candidates isolated from the screen. I am most interested in a downstream CLE signalling component that I isolated recently, a prey that the lab has long been after. In the long term, I hope to turn my projects into more publications, to be competitive in the academic job market.
And where will this work take the Hardtke lab?
CH It will force us deeper into investigating the mechanics of CLE perception. One unexpected result contributed by our collaborators, Michael Hothorn’s lab, was the observation that the CLERK ectodomain cannot interact with the ectodomains of a few known bona fide CLE receptors, in the presence of ligand. If CLERK is indeed a generic CLE co-receptor, which is a parsimonious assumption, then this indicates that something is missing to make the system work biochemically in isolation. Maybe the sensitized screen will deliver this piece of the puzzle.
Finally, let’s move outside the lab – what do you like to do in your spare time?
CH Depends on the time of year. In the winter, I take every opportunity to go skiing. Luckily, from Lausanne it only takes about one hour to reach the bottom of some of the most spectacular ski slopes on the planet. In the summer, I like to attend a random selection of sports events. In general, I like to watch people perform feats that are impossible for myself, like swimming 100 m in less than a minute…
PA …I am a mom of two young kids, 3 years and one and a half years old. I spend all my free time with them and I really enjoy it. I love to read books with them. But sometimes I also have to fold up to fit in a cardboard hut, to escape wolves or other monsters. Sometimes it is more tiring to be a mom than to be in the lab. But my favourite moments are with the kids outside, observing nature, playing with its treasures, showing them birds, insects and, of course, plants!


 (1 votes)
(1 votes)