4D Morphological Atlas of C. elegans Embryogenesis
Posted by Guoye Guan, on 2 February 2021
A joint research team co-led by Prof. Chao Tang (Peking University, PKU), Prof. Zhongying Zhao (Hong Kong Baptist University, HKBU) and Prof. Hong Yan (City University of Hong Kong, CityU) has developed a novel computational tool that can reconstruct and visualize three-dimensional shapes and temporal changes of cells, speeding up the analytical process from thousands of hours by hand to a few hours by computer. Furthermore, a total of 17 C. elegans embryos were fully segmented and normalized, generating a time-lapse 3D (i.e., 4D) morphological atlas from the 4- to 350-cell stages, including cell shape, volume, surface area, migration, nucleus position and cell-cell contact with confirmed cell identities. Both the computational tool and standard dataset can advance further studies in developmental biology, cell biology and biomechanics (Figure 1). This interdisciplinary study has been published in Nature Communications, entitled “Establishment of a morphological atlas of the Caenorhabditis elegans embryo using deep-learning-based 4D segmentation” [1].

Figure 1. The concept of 3D cell membrane imaging and segmentation (photo source: DOI number: 10.1038/s41467-020-19863-x).
The tool developed by the team is called “CShaper”. “It is a powerful computational tool that can segment and analyze cell images systematically at the single-cell level, which is much needed for the study of cell division (Figure 2), and cell and gene functions,” described Prof. Hong Yan, Chair Professor of Computer Engineering and Wong Chung Hong Professor of Data Engineering in the Department of Electrical Engineering (EE) at CityU.
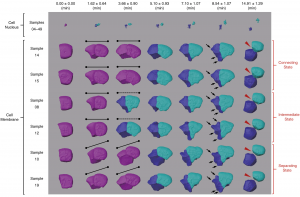
Figure 2. Morphological dynamics of cell division, deformation and migration during C. elegans embryogenesis at single-cell resolution (photo source: DOI number: 10.1038/s41467-020-19863-x).
Breakthrough in segmenting cell images automatically
Cell images are usually obtained by laser beam scanning. The existing image analysis systems can only detect cell nucleus well with a poor cell membrane image quality, hampering reconstruction of cell shapes. Also, there is a lack of reliable algorithm for the segmentation of time-lapse 3D images (i.e. 4D images) of proliferating cells. Image segmentation is a critical process in computer vision that involves dividing a visual input into segments to simplify image analysis. Often, researchers have to spend hundreds of hours labeling many cell images manually.
The breakthrough in CShaper is that it can detect cell membranes, build up cell shapes in 3D, and more importantly, automatically segment the cell images at the cell level. “Using CShaper, biologists can decipher the contents of these images within a few hours. It can characterize cell shapes and surface structures, and provide 3D views of cells at different time points (Figure 3),” said Mr Jianfeng Cao, a PhD student in Prof. Hong Yan’s lab and a co-first author of the paper.
To achieve this, the deep-learning-based model DMapNet developed by the team plays a key role in the CShaper system. “By learning to capture multiple discrete distances between image pixels, DMapNet extracts the membrane contour while considering shape information, rather than just intensity features. Therefore CShaper achieved a 95.95% accuracy of identifying the cells, which outperformed other methods substantially,” he explained. CShaper was also tested on plant tissue cells, showing promising results. The team believe the computer tool can be adopted to other biological studies.
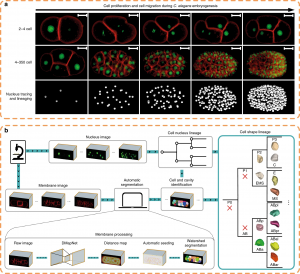
Figure 3. Fluorescent imaging (cell membrane and cell nucleus) and segmentation pipeline (CShaper) (photo source: DOI number: 10.1038/s41467-020-19863-x).
Generating a standard morphological atlas
With CShaper, the team collected 17 C. elegans embryos with fluorescence on both cell membrane and cell nucleus. The segmented regions were subjected to spatial and temporal normalization, producing a set of standardized embryo structures placed in a cuboid framework. The 4D morphological atlas lasts from the 4- to 350-cell stages during C. elegans embryogenesis, and includes quantitative information of cell shape, volume, surface area, migration, nucleus position and cell-cell contact with confirmed cell identities (Figure 4). “The morphological atlas at cellular level is information-rich and allows systematic analysis with cell identity, and is needed by the field of developmental biology,” said Prof. Zhongying Zhao.
“With such accurate data, we can not only develop mathematical model to ‘compute’ embryogenesis, but also search the underlying principles which govern the nematode development,” said Prof. Chao Tang. “Using the standard morphological data, recently we built a phase field model to reconstruct the C. elegans embryonic morphogenetic procedure [2], and uncovered a minimal time principle optimizing the C. elegans embryonic cell lineage [3],” said Mr Guoye Guan, a PhD student in Prof. Chao Tang’s lab and a co-first author of the paper.
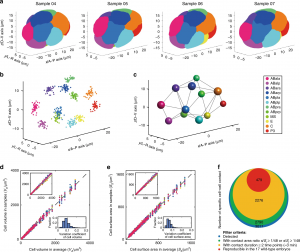
Figure 4. Standard morphological atlas and multidimensional developmental properties (photo source: DOI number: 10.1038/s41467-020-19863-x).
Prof. Chao Tang (PKU), Prof. Zhongying Zhao (HKBU) and Prof. Hong Yan (CityU) are the corresponding authors of the paper; Ph.D students Jianfeng Cao (CityU) and Guoye Guan (PKU) and Dr. Vincy Wing Sze Ho (HKBU) are the co-first authors; Ming-Kin Wong (HKBU) and Lu-Yan Chan (HKBU) provided experimental assistance.
The funding support for this study included the Hong Kong Research Grants Council, the HKBU Interdisciplinary Research Cluster Fund, the Hong Kong Innovation and Technology Commission, the Hong Kong Institute for Data Science, the Ministry of Science and Technology of China, and the National Natural Science Foundation of China.
Part of this article is from the news release in PKU [4] and CityU [5].
References
[1] Jianfeng Cao, Guoye Guan, Vincy Wing Sze Ho, Ming-Kin Wong, Lu-Yan Chan, Chao Tang, Zhongying Zhao, Hong Yan. Establishment of a morphological atlas of the Caenorhabditis elegans embryo using deep-learning-based 4D segmentation. Nature Communications, 2020, 11: 6254 (https://www.nature.com/articles/s41467-020-19863-x).
[2] Xiangyu Kuang, Guoye Guan, Ming-Kin Wong, Lu-Yan Chan, Zhongying Zhao, Chao Tang, Lei Zhang. Computable early C. elegans embryo with a data-driven phase field model. bioRxiv, 2020, 422560 (https://www.biorxiv.org/content/10.1101/2020.12.13.422560v2).
[3] Guoye Guan, Ming-Kin Wong, Zhongying Zhao, Lei-Han Tang, Chao Tang. Lineage optimization in nematode’s early embryogenesis. arXiv, 2020, 2007.05723 (https://arxiv.org/abs/2007.05723).
[4] Standard morphological atlas of C. elegans embryo (http://cqb.pku.edu.cn/tanglab/News/shownews.php?id=224&lang=en).
[5] A powerful computational tool for efficient analysis of cell division 4D image data (https://www.cityu.edu.hk/research/stories/2020/12/22/powerful-computational-tool-efficient-analysis-cell-division-4d-image-data).


 (5 votes)
(5 votes)