The people behind the papers – Awais Javed and Michel Cayouette
Posted by the Node Interviews, on 15 January 2021
This interview, the 88th in our series, was published in Development last year.
The mammalian retina contains a variety of functionally distinct cell types that are generated by progenitor cells in a specific chronological order. A new paper in Development probes the role of the POU-homeodomain factors Pou2f1 and Pou2f2 in the timely generation of cone photoreceptors in mice. We caught up with first author and PhD student Awais Javed and his supervisor Michel Cayouette (Director of the Cellular Neurobiology Research Unit at the Montreal Clinical Research Institute, Professor at the Université de Montréal and Adjunct Professor at McGill University) to hear more about their work.

Michel, can you give us your scientific biography and the questions your lab is trying to answer?
MC: I obtained my PhD in Neurobiology from Université Laval, in Québec city, Canada, working on viral vector-mediated gene transfer approaches in mouse models of retinal degeneration. Towards the end of my PhD I became very interested in understanding how all the beautiful cell types I was looking at under the microscope were generated, and decided I would study neural development during my postdoc. I contacted several labs and, fortunately for me, the famed developmental neurobiologist Martin Raff, whom I admired greatly, offered me the last postdoc spot available in his lab before he retired. Realizing this unique opportunity, my wife and I decided to move to London where I spent 3 years studying asymmetric cell divisions in retinal progenitors and the relative contribution of intrinsic and extrinsic signals in cell fate specification. When Martin retired, I was in the middle of a project that I wanted to finish before looking for an independent position. I then joined Ben Barres’ lab at Stanford University, USA, where I continued to work for 2 years on the project, which was finally published in 2003. In November 2004, I started my independent career in Montreal.
Ever since the beginning, my lab has been focused on studying how cell diversification is achieved during nervous system development, with the long-term goal of using this knowledge as the basis to develop regenerative therapies. We are particularly interested in understanding how neural progenitors know when it is time to make a specific combination of cell types and, once a temporal window has been defined, how progenitors choose between alternative cell fates available to them at that time. We primarily use the mouse retina as a model system to address these questions, but we have also studied myelination and the inner ear to ask questions related to cell polarity, which we also study in the context of asymmetric cell division in the retina.
And Awais – how did you come to work in Michel’s lab and what drives your research today?
AJ: In the final year of my undergraduate degree at University College London, I was revising the course material for the end of term developmental biology exam, including the temporal competence cascade in neuroblast ventral nerve cord specification in Drosophila. I found it fascinating how the same set of genes could regulate cell fate in different parts of the ventral nerve cord, and I wondered if there was a similar cascade in the vertebrate central nervous system. A quick PubMed search led me to the work of the Cayouette lab, who showed that Ikaros, the fly hunchback homolog, was a temporal competence factor in the mammalian retina. As it so happens, I was planning to move to Canada for personal reasons and the first lab I looked up was Michel’s. I started my PhD trying to find similarities between vertebrate and invertebrate neurodevelopment, but now I feel more driven by the complexity biology has to offer and how different the systems can be.
How has your research been affected by the COVID-19 pandemic?
AJ: I was very fortunate because on the last day before the lockdown; we received the next-gen sequencing results for a number of experiments I had submitted in February. The first 2 months of the shutdown, I learned how to analyse these datasets using R and Python. I had no previous knowledge of using any programming language, so it was a true test of perseverance. When the lab finally opened up, I was just happy to be back at the bench: I didn’t think I would ever say this but I definitely missed genotyping!
I didn’t think I would ever say this but I definitely missed genotyping!
MC: The lab has been completely shutdown with only essential activities allowed for almost 2 months. Like Awais, people in the lab tried to use this difficult period to make some progress. Some wrote a draft of their paper, analysed large datasets or counted cells, while others took online classes and learned how to code. I was very proud of my group – they were amazing and really used this time as best they could, despite all the challenges associated with the situation. But of course, everything was delayed in the lab. Most difficult for us was having to scale down our animal colony, as the animal facility staff was reduced. We are just now getting back to normal after several months. This also meant it took longer than expected to carry out the revisions for our paper, but the Development editors were helpful in guiding us to prioritize experiments and were flexible with the time allowed for us to do these, which was appreciated!
Why is timing critical for the generation of cellular diversity in the retina?
AJ & MC: While it is not always clear why specific neurons must be generated before others, it is generally accepted that cell birth order is critical to ensure proper neural circuit formation. Just as the foundation of a house must be built first because other parts of the house sit on it, certain types of neurons constitute the foundation of a given circuit, as they receive inputs from neurons produced later. This tightly regulated chronology is critical for the generation of highly complex tissues.
Can you give us the key results of the paper in a paragraph?
AJ & MC: In this paper, we investigated the role Pou2f1 and Pou2f2 in the developing mouse retina. We show that both genes are necessary and sufficient for cone photoreceptor cell fate specification during retinal development. We further report that Ikzf1, an early temporal identity gene that we previous identified in the retina, upregulates Pou2f1, which in turn represses the late temporal identity factor Casz1, thereby defining a temporal identity window conducive to cone photoreceptor production. Mechanistically, we show that Pou2f1 activates Pou2f2, which then represses expression of the rod-promoting factor Nrl in postmitotic photoreceptor precursors by binding to a POU-specific site in the promoter, thereby favouring the cone fate. As Pou2f1 and Pou2f2 are orthologues of fly pdm, which is well known for its role in neuroblast temporal patterning, our results, together with previously published studies, suggest that some aspects of this cascade are conserved in vertebrates. This work also establishes a link between temporal identity genes and cell fate determinants in the mammalian central nervous system.
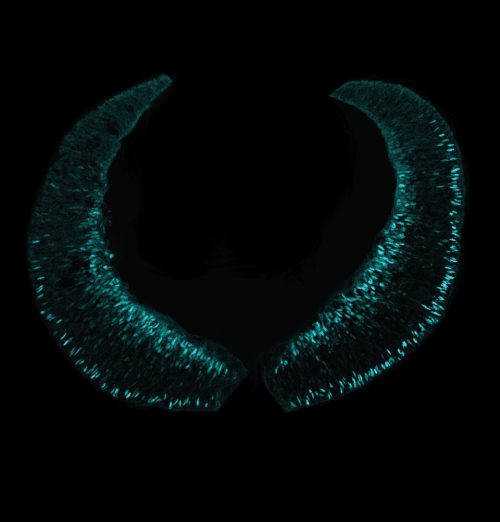
Loss of Pou2f2 reduces, but does not eliminate, cone production: how are cones determined in its absence?
AJ & MC: It is of course possible that other genes compensate for the loss of Pou2f2. Potential candidates include Onecut1 and Onecut2, as a partial loss of cones, similar to what we observed in this study, is observed in double knockout animals. Another possibility is that abolishing the temporal window for cone production induces progenitors to generate cones outside the normal window using alternative pathways. Finally, we have previously suggested that loss of temporal identity in progenitors does not lead to complete absence of any given cell type, but simply to a reduced probability of generating these cell types, which might explain why we do not observe a complete loss of cones in this study.
Do you know what restricts Pou2f1 expression to early RPCs? Is there evidence of mutual inhibition with other temporal factors?
AJ & MC: This is a very interesting question. We show in our paper that Ikzf1 upregulates Pou2f1 expression in early RPCs, but it remains unknown what actually turns off Pou2f1 expression in later progenitors. An obvious candidate is Casz1, which might repress Pou2f1 at later stages, similar to what is observed in Drosophila neuroblasts, but our preliminary experiments looking at this possibility do not appear to support this model. Another possible regulator of Pou2f1 is Foxn4, which was recently discovered as a regulator of Ikzf1 and Casz1 in the mammalian retina. More work is needed to fully elucidate this issue.
When doing the research, did you have any particular result or eureka moment that has stuck with you?
AJ: The main result that comes to mind was actually one of the first observations that led us down the path of this paper. Initially, immunostainings of Pou2f1 showed strong expression in ganglion cells, as well as other cell types, which we later found out to be cone and horizontal cells. When we first tried gain-of-function experiments with Pou2f1 using a retroviral vector, we did not initially consider that cone production would be induced. Out of sheer curiosity, I added S-opsin as a marker for cones and, to my surprise, I found S-opsin+ cells in the overexpression condition. My first thought was that I had mixed up the viruses or the immunostainings, but when it started to repeat, I was convinced it was a real result. It wasn’t a eureka moment per se, but it felt quite good to stumble onto a finding that would later turn out to be the central hypothesis of the paper.
And what about the flipside: any moments of frustration or despair?
AJ: The ChIP-qPCR experiments, without a doubt. It is running joke in the lab at this point because of how tired I was during lab meetings when I was working on those experiments. It was very labour intensive because I used embryonic retinal tissues and I had quite a bit of optimization to do before I could get any results. But it was well worth it, when I finally got the experiment to work. In the end, it was a fantastic lesson learned on perseverance.
What next for you after this paper?
AJ: I am finishing up a few projects in the lab and hoping to graduate in early 2021. I am currently looking for postdoc opportunities and I am very excited to continue investigating cell fate specification in other parts of the central nervous system.
Where will this story take the Cayouette lab?
MC: We are becoming increasingly interested to determine whether temporal factors could be used to promote tissue regeneration. The idea is simple, as early temporal factors are sufficient to reprogram late-stage progenitors into generating early-born cell types, we wonder whether these factors might also be able to reprogram differentiated cells into neural progenitors. We would also like to determine whether progenitors invariably go through all the temporal identity windows defined by the temporal factors we have studied so far in the retina (Ikzf1, Pou2f1, Pou2f2 and Casz1) or whether some can skip a given window. Answers to this question might explain the huge heterogeneity of clonal composition observed in lineage-tracing studies in the retina. This will require the development of tools to follow temporal factor expression in real time in single cells, but I think it would be very cool to address this question, as it has far-reaching implications. Finally, detailed mechanistic understanding of temporal patterning remains poorly understood, even in flies. We are becoming increasingly interested in this question. It is likely that temporal identity is defined by specific epigenetic landscapes, and whether and how temporal factors shape chromatin conformation and nuclear architecture is a problem we will likely focus on in years to come.
Finally, let’s move outside the lab – what do you like to do in your spare time in Montreal?
AJ: I am an avid painter and it has helped me deal with the ups and downs of a PhD. I also founded a drama club at the institute. We wrote and directed a couple of plays to a full audience at one of the institute’s auditoriums, which was an amazing experience. I did not realize that many scientists are fantastic artists!
MC: Since I was a child, I have been playing ice hockey and I reached fairly high competitive level. To this day, I continue to play once or twice a week, often with students who seem to be getting younger and faster each year… I am also a fervent cyclist and love to ride the roads of the countryside and the trails of the provincial park around our house. Finally, I love great food and wine, and since Montreal has a large choice of fantastic restaurants, it is a good place to be!


 (No Ratings Yet)
(No Ratings Yet)