Moving in concert: How lateral line primordium cells coordinate to migrate
Posted by ttcolak, on 21 September 2019
Written by Tugba Colak-Champollion
Story behind our recent paper in Current Biology “Cadherin-Mediated Cell Coupling Coordinates Chemokine Sensing across Collectively Migrating Cells” (Tugba Colak-Champollion, Ling Lan, Alisha R. Jadhav, Naoya Yamaguchi, Gayatri Venkiteswaran, Heta Patel, Michael Cammer, Martin Meier-Schellersheim, Holger Knaut)
Guided cell migration is a crucial event in many biological and mechanical processes. During development, orchestrated movements of cells pattern tissues and organs. Wounds in our bodies close by the migration of cell sheets. Cell migration is one of the weapons of the immune system, which sends leukocytes to the site of infection to fight against pathogens. But cell migration also contributes to several pathological conditions, such as the dissemination of cancer cells to the sites of metastasis in the body. Thus, detailed studies of mechanisms of cell migration are likely to improve our understanding of animal development, homeostasis, and disease progression.
Cell migration occurs in two major modes: single cell migration and collective cell migration. Single cell migration can be defined as a cell migrating on its own upon extracting directional information and polarizing and moving towards the target site. While collectively migrating cells also extract directional information and polarize toward the target site, they also have the additional task of coordinating with each other to move in the same direction and at the same speed.
How do cells move as a coherent collective following guidance cues? In the latest study from our lab [1], my coauthors and I investigated how directional sensing in collectively migrating cells is organized. We used the Zebrafish lateral line primordium (primordium) as a model for our study. The primordium is a collective of tightly adhering ~100 cells, which originates behind the ear of the fish at around 18 hours post fertilization (hpf) and migrates to the tip of the tail by 42 hpf.
The primordium migration is guided by chemokine signaling. For proper migration, the primordium requires the chemokine Cxcl12a and its receptors Cxcr4b and Cxcr7b. Importantly, the signaling receptor Cxcr4b is expressed in every primordium cell, while the Cxcr7b expression is restricted to the rear of the collective, where it acts as a sink receptor and generates a Cxcl12a gradient across the tissue [2-5].
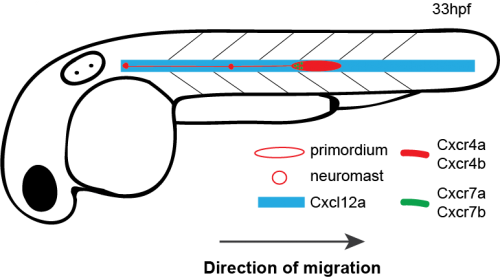
Schematic of primordium migration and chemokine signaling system in primordium
Primordium migration in wild-type embryo
In collective cell migration, a prevailing model suggests that there is a division of labor between two groups of cells: leaders and followers. These two groups of cells have both topological and functional attributes. For example, leaders are located in front of the collective, whereas followers are located in the rear. In terms of functionality, the model suggests that leader cells explore their surroundings extracting directional information and relaying this information to the follower cells.
It was proposed that the primordium migration also follows a leader-follower model. An elegant study by Haas and Gilmour generated supporting evidence for this model using chimeric analysis [6]. They placed wild-type cells, which can see the chemokine signal into cxcr4b mutant primordia whose migration stalls prematurely. When at least a few wild-type cells ended up in the front region, the migration was restored – albeit at a slower speed than the speed of wild-type primordia.
However, when I started my Ph.D. project in the Knaut lab, two studies that recently were published made my advisor Holger Knaut and me rethink the leader-follower model [4, 5]. These two studies demonstrated that there is a linear chemokine signaling gradient across the tissue that is essential for the directionality of the primordium. This gradient is available to both leader and follower cells, not just to the leader cells.
Meanwhile, Gayatri Venkiteswaran, a post-doc in our lab, made an interesting observation when she was scoring the primordium migration phenotype in chemokine receptor mutants. Previously, it was known that loss of Cxcr4b leads to primordium stalling. However, loss of the other Cxcr4 receptor, Cxcr4a, that is also expressed in the primordium cells [7] does not result in a primordium migration defect. Gayatri found that taking away one copy of cxcr4a from cxcr4b-/-primordia makes the cxcr4b-/- phenotype worse and taking away both copies of both receptors is much worse than the cxcr4b-/- or cxcr4a+/-; cxcr4b-/-phenotypes. This observation suggested that Cxcr4a also contributes to migration and a primordium that is cxcr4b-/- can still see a little bit of the chemokine signal. This meant the earlier study which suggested that the existence of a few chemokine-sensitive cells could restore the migration phenotype of chemokine-blind (cxcr4b-/-) cells was conducted using primordia that could still see some of the directional signal because they retained functional Cxcr4a.
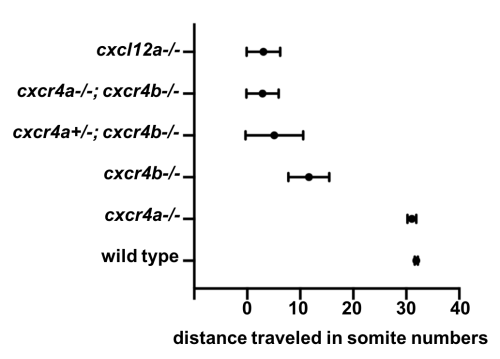
Quantification of primordium migration in 48 hpf embryos of indicated genotypes
Primordium migration in cxcr4b-/- embryo
Primordium migration in cxcr4a-/-; cxcr4b-/- embryo
In the light of these two pieces of new information—existence of a linear gradient across tissue and Cxcr4a’s contribution to directional migration—we decided to take a closer look at the leader-follower model and the contribution of follower cells to directionality and speed of the primordium.
Our first question was whether a few chemokine-sensitive (wild-type) cells could restore the migration phenotype of completely blind (cxcr4a-/-; cxcr4b-/-) primordia. To answer this question, we used a classical developmental biology technique named chimeric analysis. The idea was putting cells from wild-type donor embryos into cxcr4a-/-; cxcr4b-/- host embryos at the blastula stage. The donor primordium cells are labeled with a red transgenic marker and the host primordium cells are labeled with a green transgenic marker. After transplantation, the embryos are grown until 30 hpf and the transplanted host embryos are screened for the presence of red donor cells in the primordium. Although chimeric analysis provides very clean and reliable data, it is an inefficient technique. Unfortunately, not every single embryo transplanted gets the donor cells in the primordium. Additionally, it is a difficult technique and takes a while to perfect it. Even when you become very good at it, you can still have some days when the host mortality is high for unknown reasons.
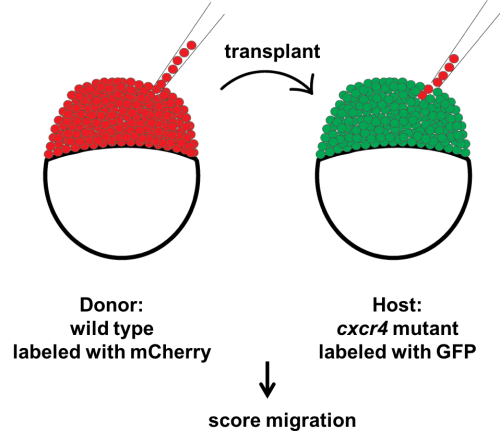
Schematic of blastomere transplantation
Despite the difficulty and inefficiency of chimeric analysis, we still chose to do it because it was the best way to answer our questions. There was an additional layer of difficulty which turned out to be a good thing later: to generate cxcr4a-/-; cxcr4b-/-embryos, we had to in-cross cxcr4a+/-; cxcr4b-/- fish because cxcr4a-/-; cxcr4b-/- adults are not viable. This meant only a quarter of our host embryos would have the desired genotype. Half would be cxcr4a+/-; cxcr4b-/- and a quarter would be cxcr4b-/-. We had no way of knowing the genotype of the chimeric host embryos until going through a genotyping protocol after imaging these embryos. Nevertheless, we got what we wanted and more.
We found that a few chemokine sensitive (wild-type) cells do not restore the migration phenotype of completely blind (cxcr4a-/-; cxcr4b-/-) primordia, an observation that is inconsistent with the classical leader-follower model. However, we had one embryo in which about half of the primordium consisted of wild-type cells, and the other half consisted of cxcr4a-/-; cxcr4b-/-cells, and this chimeric primordium migrated about half of its path. This made us consider the possibility that cells pull on each other during migration. When there are many cells which can see the chemokine, they might generate sufficient pulling forces in the right direction to drag their chemokine blind neighbors along—in the case of this chimeric embryo, half of the way.
We decided to follow up on this observation using a quantitative approach. We predicted that as we increase the number of chemokine-sensing cells in an otherwise chemokine-blind primordium, the distance migrated by the chimeric primordium should increase. To quantify this behavior, we needed a lot of samples. Using chemokine-blind primordia (cxcr4a-/-; cxcr4b-/-) as hosts posed two challenges. First, it is difficult to get them (remember that only one-quarter of the embryos are of this genotype). Second, it might require a lot of chemokine-sensing cells to pull their completely chemokine-blind neighbors along. To overcome these challenges, we considered using cxcr4a+/-; cxcr4b-/- primordia as hosts. These almost chemokine-insensitive primordia have only one copy of cxcr4a left. Gayatri’s quantification of the migration behavior showed that the migration of these cxcr4a+/-; cxcr4b-/- primordia are worse than cxcr4b-/-primordia and a little bit better than cxcr4a-/-; cxcr4b-/- primordia. Having just a little bit of Cxcr4a activity would help us resolve the relationship between migrated distance and chemokine-sensing cell number, we hoped.
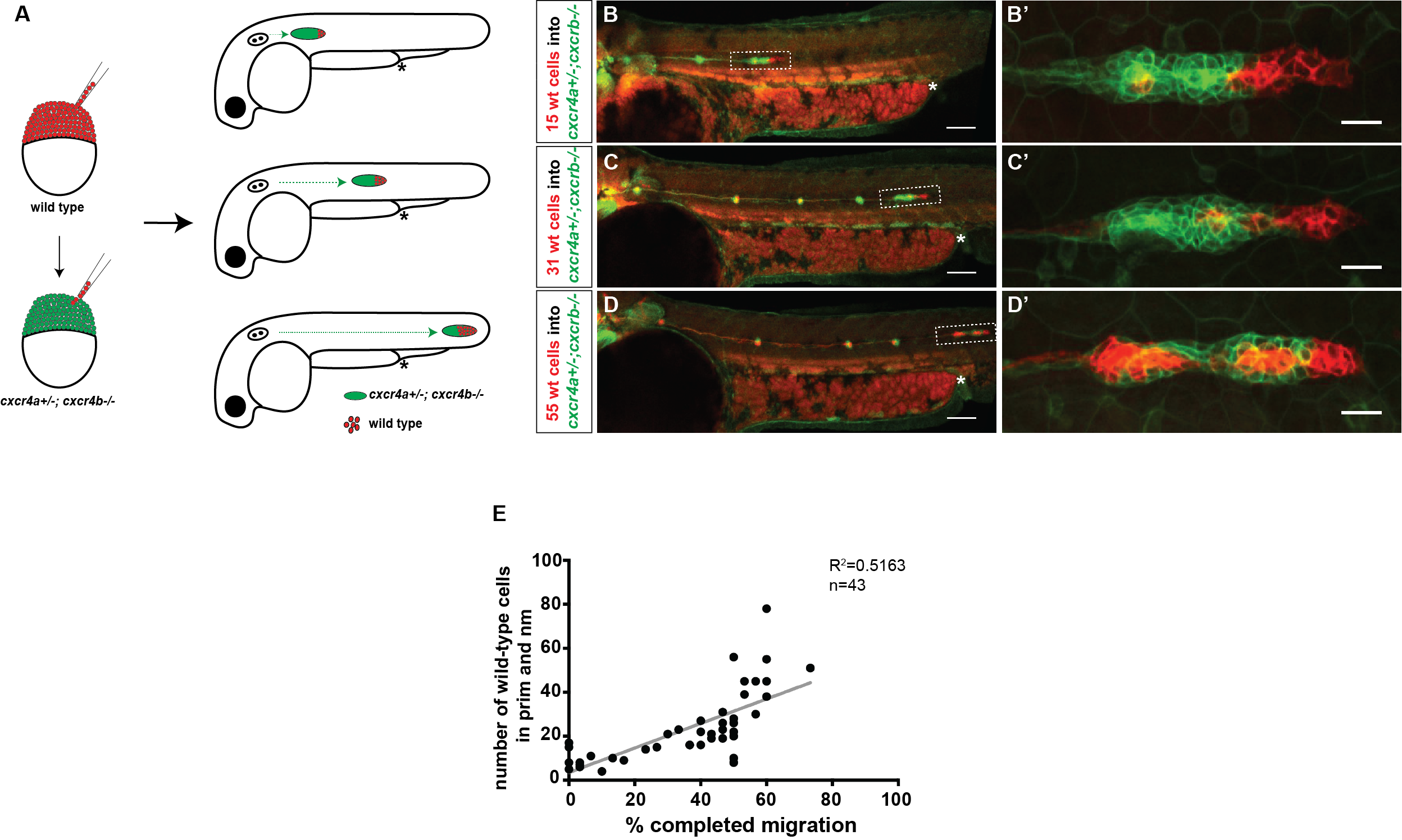
A) Schematic of experimental design and predictions. B-D’) Examples of chimeric primordia with low, medium, and high donor cell contribution. E) Graph of the total number of wild-type cells in the chimeric primordia (total primordial wild-type cells include interneuromast, neuromast (nm) and primordium (prim) cells) versus the migration distance of cxcr4-deficient primordia. 48 hpf.
Luckily, we already imaged a lot of chimeric primordia that consisted of chemokine-sensitive (wild-type) and chemokine-insensitive (cxcr4a+/-; cxcr4b-/-) cells. So we went back to this data set and counted the number of wild-type cells and measured how far these chimeric primordia migrated. As we predicted, the chimeric primordia migrated further as the number of chemokine-sensitive cells increased. This observation was consistent with the idea of cells “pulling” on each other. Next, we asked what could facilitate this “pulling”. We turned to the obvious candidates: cell-cell adhesion molecules. And there are a few of them expressed by the primordium cells including E-cadherin (Cadherin 1, Cdh1) and N-cadherin (Cadherin 2, Cdh2) [8-10].
We first decided to focus on Cdh1 and Cdh2. Our first question was how cdh1-/- and cdh2-/- cells behave during migration. The simplest method of answering this question would be observing the migration behavior of cdh1 or cdh2 mutant primordia under a microscope. However, this was not an option for either of these two genotypes. The problem with cdh1-/- embryos is that they die during gastrulation before the primordium is formed. As for cdh2-/- embryos, they have problems with somite development that affect the proper expression of Cxcl12a (the directional cue). Thus, we went back to our favorite technique, chimeric analysis by blastomere transplantation.
Live imaging of cdh1-/- or cdh2-/- cells in an otherwise wild-type primordium showed that lacking either of the cadherins does not affect migration. These mutant cells co-migrated normally with their wild-type neighbors. This raised the next question: Are these cadherins acting redundantly? To answer this question, we needed to generate cdh1-/-; cdh2-/- embryos to be used as donors. However, this is a real challenge, as the cadherin mutants are not adult viable. To obtain the desired genotype, we had to in-cross cdh1+/-; cdh2+/- fish. The chance of finding a double cadherin mutant embryo is one in sixteen! Despite the odds, we did this experiment—repeatedly. We never found a chimeric embryo whose donor embryo was cdh1-/-; cdh2-/-. But we obtained a good sample size that made us confident to think that either such double cadherin mutant cells do not become primordium cells or they dissociate from the primordium very early on. Despite not being able to find what we set out to find, we encountered some interesting genetic scenarios. For example, cdh1-/-; cdh2+/- cells dissociated from the migrating primordium when they were placed at the tip of the primordium, whereas cdh1+/-; cdh2-/- cells fell off the primordium when they were located at the rear of the primordium. These findings were puzzling until the next observation.
To observe Cdh1 and Cdh2 expression in the primordium, we made BAC transgenic lines that expressed Cdh1-GFP and Cdh2-mCherry. A closer look at the primordia in these transgenic fish revealed that Cdh1 was expressed more in the front region of the primordium and Cdh2 was expressed more in the rear. This could be a possible explanation for the observation mentioned above, that cdh1-/-; cdh2+/- cells fell off from the tip positions and cdh1+/-; cdh2-/- cells fell off from the rear positions. Perhaps Cdh1 was needed more in the front and Cdh2 more in the rear?
The observation of cadherin deficient cells falling off from the primordium also suggested that cell-cell adhesion through cadherins couples the migrating cells. However, is such cell coupling necessary for cells to pull on each other? To answer this question, we transplanted cdh1-/- or cdh2-/- cells into cxcr4b-/-primordia. While cxcr4b-/- primordia have a migration defect due to not being able to see the chemokine signal properly, cadherin mutant cells still have the chemokine receptors and should migrate directionally. To our surprise, cdh1-/- cells located in the front region split away from the cxcr4b-/- cells. But cdh2-/- cells located in the front region pulled the cxcr4b-/- cells along. This finding was consistent with the differential expression pattern of Cdh1 (more in front) and Cdh2 (more in rear) that we had observed earlier. Additionally, a little bit of literature digging revealed that in vitro studies suggested that Cdh1 could withhold more force than Cdh2 [for example 11]. It is plausible that there is increased tension between the donor population, which moves persistently in a specific direction, and the host population, which moves in random directions. A strong physical attachment might be necessary to keep these two groups together.
Wild-type donor cells (red) partially restore migration defect of cxcr4b-/- primordium (green)
cdh1-/- donor cells (red) split away from cxcr4b-/- host primordium (green)
cdh2-/- donor cells (red) partially restore migration defect of cxcr4b-/- primordium (green)
These findings suggested that the physical coupling of primordium cells is important for the group’s directional migration. But how does cell-cell adhesion affect individual cell directionality within a group? Up on discussions of our data with Martin Meier-Schellersheim, who is a physicist at the NIH and a co-author, we decided to take a quantitative approach using nuclear movement as a proxy for cell movement.
Next, we utilized our favorite technique, blastomere transplantation, once again and placed donor cells (wild type, cdh1-/-, cdh2-/-, or cxcr4b-/-) labeled with H2A-GFP into wild-type host primordia whose cell nuclei were labeled with H2A-mCherry.
Using commercial image software, we tracked the individual donor and host nuclei in different primordium locations at high spatial and temporal resolution. To analyze the tracking data, we used custom scripts to assess directional sensing using three measures: neighbor-neighbor distance, directionality index, and directionality angle. The nuclear tracking analysis showed that directional sensing of cadherin and cxcr4b deficient cells was impaired based on all three categories, lack of Cdh1 having the most severe effect. Together these data showed that in addition to directional cue sensing, efficient migration requires cadherin-mediated cell coupling.
Our observations suggested that cadherin-mediated cell-cell adhesion is important for coordinating cell movements in the migrating primordium. To test this further, we decided to use a gene trap line that expresses functional alpha E-catenin tagged with Citrine (Ctnna1-Citrine) from the endogenous promoter that we recently obtained from Scott Fraser’s lab [12]. Ctnna connects Cadherins on the plasma membrane to the actin cytoskeleton; therefore, lack of Ctnna should impair cadherin-mediated cell-cell adhesion. Chimeric analysis using blastomere transplantation would be the way to approach this experiment since the ctnna-/- embryos die during somitogenesis, before the primordium develops. But luckily, our lab recently developed a protein degradation system named zGrad that could be an efficient alternative to chimeric analysis [13]. As expected, the time-lapse analysis showed that depletion of Ctnna1 resulted in primordia that migrate less directionally. Interestingly, cells separated from each other during migration forming large irregular gaps consistent with the idea that cadherin-catenin complexes mediate the adhesion among the cells in the primordium.
In summary, these results suggest that all the cells in the primordium interpret the directional information and are physically coupled to each other to achieve robust migration. This behavior is not unlike some Turkish folk dances, which are characterized by groups of dancers who hold hands tightly as they dance to a tune in a synchronized fashion. Just as each dancer needs to listen to the music, each cell needs to sense the directional signal in order to coordinate their movements. Through their tight connections, dancers and cells alike synchronize their individual motions, thereby moving in unison.
References
- Colak-Champollion, T., et al., Cadherin-Mediated Cell Coupling Coordinates Chemokine Sensing across Collectively Migrating Cells.Curr Biol, 2019. 29(15): p. 2570-2579 e7.
- Dambly-Chaudiere, C., N. Cubedo, and A. Ghysen, Control of cell migration in the development of the posterior lateral line: antagonistic interactions between the chemokine receptors CXCR4 and CXCR7/RDC1.BMC Dev Biol, 2007. 7: p. 23.
- Valentin, G., P. Haas, and D. Gilmour, The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b.Curr Biol, 2007. 17(12): p. 1026-31.
- Dona, E., et al., Directional tissue migration through a self-generated chemokine gradient.Nature, 2013. 503(7475): p. 285-9.
- Venkiteswaran, G., et al., Generation and dynamics of an endogenous, self-generated signaling gradient across a migrating tissue.Cell, 2013. 155(3): p. 674-87.
- Haas, P. and D. Gilmour, Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line.Dev Cell, 2006. 10(5): p. 673-80.
- Siekmann, A.F., et al., Chemokine signaling guides regional patterning of the first embryonic artery.Genes Dev, 2009. 23(19): p. 2272-7.
- Matsuda, M. and A.B. Chitnis, Atoh1a expression must be restricted by Notch signaling for effective morphogenesis of the posterior lateral line primordium in zebrafish.Development, 2010. 137(20): p. 3477-87.
- Revenu, C., et al., Quantitative cell polarity imaging defines leader-to-follower transitions during collective migration and the key role of microtubule-dependent adherens junction formation.Development, 2014. 141(6): p. 1282-91.
- Kozlovskaja-Gumbriene, A., et al., Proliferation-independent regulation of organ size by Fgf/Notch signaling.Elife, 2017. 6.
- Panorchan, P., et al., Single-molecule analysis of cadherin-mediated cell-cell adhesion.J Cell Sci, 2006. 119(Pt 1): p. 66-74.
- Trinh le, A., et al., A versatile gene trap to visualize and interrogate the function of the vertebrate proteome.Genes Dev, 2011. 25(21): p. 2306-20.
- Yamaguchi, N., T. Colak-Champollion, and H. Knaut, zGrad is a nanobody-based degron system that inactivates proteins in zebrafish.Elife, 2019. 8.


 (8 votes)
(8 votes)