Introducing ACME: the species-versatile fixation and dissociation solution for single cell analysis
Posted by nathankenny, on 1 July 2020
This post highlights the approach and findings of a new research article available in preprint on BioRxiv. This feature was written by members of the Solana lab, authors of that paper.
Single cell techniques are revolutionising biology, but at the moment they are largely limited to traditional model organisms and require access to specialised equipment in traditional laboratory settings. We are excited to introduce ACME, a novel technique which promises to democratise access to the world of single cell analysis, simplifying the extraction and processing of samples. We think this will be a ground-breaking innovation for cell biology, developmental biology and evo devo research.
ACME (when not written on an anvil in a Warner Bros cartoon), stands for ACetic acid-MEthanol dissociation. This principle stems from a 19th century approach for cell dissociation for microscopy. For instance, see its application in Hydra and in planarians. We show that, with modification, this same principle works for single cell transcriptomics, profiling the cellular atlases of two planarian species using SPLiT-seq.
What are the benefits of ACME dissociation?
An ongoing problem in single cell sequencing analysis is the time taken between sampling and fixing cells. During this time, cells are alive and under stress, which causes changes to their transcription and cellular identity. Nuclei based methods can get around this problem to an extent, but yield much lower RNA content and have their own technical requirements.
ACME cells are fixed from the time of dissociation and can be cryopreserved one or several times at several points throughout the process. This can be right after dissociation, for instance, in the field or when doing multi step protocols. In the lab, this can also be after FACS enrichment. This will allow exchange of samples between labs, preservation of fixed patient material, and freezing large sample sets for parallel analysis (avoiding some batch effects). RNA from ACME dissociated cells has high integrity, provided that RNAse-free conditions are met. Even after 5 cycles of freezing and thawing we recover RNAs with acceptable integrity – but one or two freezing steps should be enough for most applications.
ACME disassociation also preserves cell morphologies – this could be a game-changer by allowing a variety of integrated approaches to studying cellular evolution and homologies, which are not possible with most current cell fixation methods for scRNAseq.
In short, ACME greatly streamlines the preparation of single cells for scRNAseq, while allowing approaches to answer questions beyond the scope of current methods.
How is ACME dissociation performed?
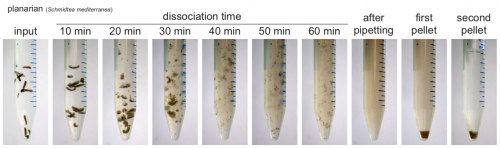
ACME solution is made from common lab reagents, including (unsurprisingly) acetic acid and methanol. We show that this method efficaciously dissociates planarians in ~1h. Cells are then resuspended in PBS with BSA for molecular biology. We have successfully used ACME dissociation with animals including zebrafish, fruit flies, spiders, annelids, snails and sea anemones. Sometimes the harder outer layers of animals or embryos must be removed, but this is usually a simple process, and normally can be adapted from standard protocols.
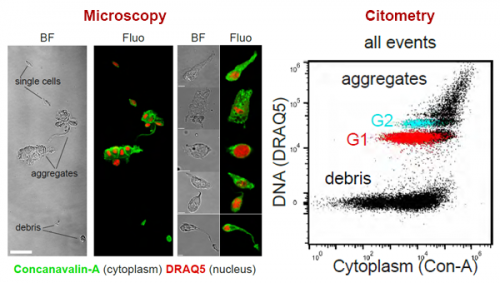
We demonstrate a simple staining process for flow cytometry and FACS. This is based on a nuclear stain (DRAQ5) and a cytoplasmic stain (Concanavalin-A). This allows us to distinguish ACME dissociated single cells from aggregates and debris.
Test-driving ACME
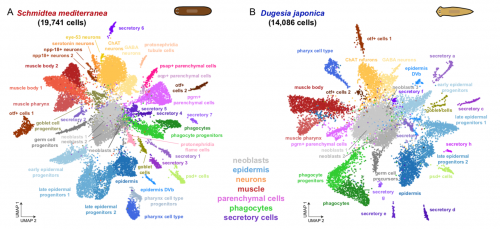
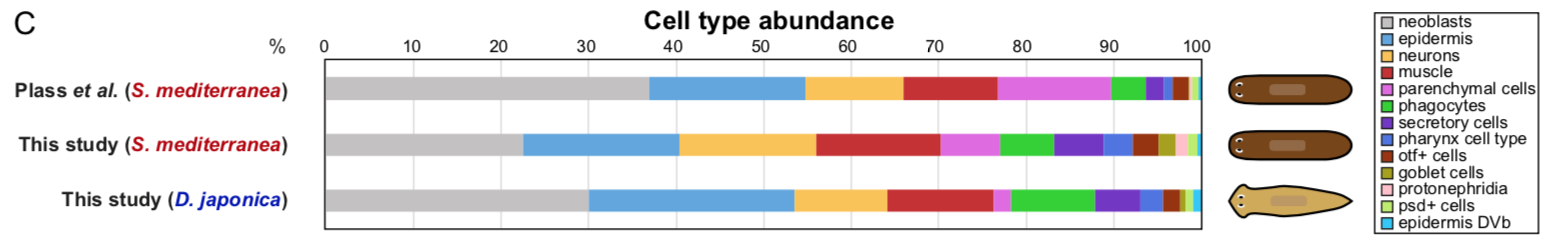
To test that ACME dissociated cells can be used for combinatorial barcoding scRNAseq we performed SPLiT-seq in a species mixing experiment, including two cryopreservation steps. We managed to sequence 34K cells from the two species, with excellent species separation. Our models were the planarian Schmidtea mediterranea (which has been subject to scRNAseq before, allowing us to benchmark our results) and a second planarian species, Dugesia japonica (which hasn’t been sequenced, allowing us to prove that ACME copes with novel species just as well!).

In the planarian Schmidtea mediterranea we find markers of all previously described cell types, showing that ACME dissociation does not artificially deplete any known kind of cell. As a bonus, we find some new clusters, such as the nanos+ germ cell progenitors, which were known to exist, but had not been noted as distinct clusters in planarian scRNA seq data before. The early fixation provided by ACME potentially allowed these cells to be fixed quickly, while it is possible they lost their distinct expression profile in previous experiments.
We also describe for the first time the cell type atlas of a second planarian species, Dugesia japonica. This shows many initial similarities with Schmidtea mediterranea – the most abundant cell type groups in both species are epidermal, neural and muscular cells, present at comparable proportions. The depth of sampling, however, will allow us to compare even rarer cell type abundance and gene expression, allowing species vs species comparison at excellent resolution, even off a single cell sequencing run.
Future possibilities, and please try it out!
We plan to use ACME to explore cell type evolution in a variety of non-model organisms, However, it could also be of broad use in more traditional model organisms, particularly when fixing multiple samples for simultaneous analysis. While we use SPLiTseq, there is no conceptual reason why it would not also work for droplet-based techniques, and could be adapted to solve many problems in current single cell sequencing protocols. Further optimisation will of course continue to take place, and we will be happy to offer advice to anyone willing to try it in their own odd organism.
In short, ACME is a versatile and powerful cell dissociation method for single-cell transcriptomics, providing early fixation and easy storage of material. This will greatly enhance investigations of cell type diversity and dynamics in multiple different organisms – particularly in challenging conditions.
This work was lead by Helena García-Castro and Jordi Solana, with Nathan Kenny, Patricia Álvarez-Campos and Vincent Mason from the Solana Lab, Oxford Brookes, and collaborators from across the UK and Europe (Anna Schonauer, Vicky Sleight, Jakke Neiro, Aziz Aboobaker, Jon Permanyer, Marta Iglesias, Manuel Irimia and Arnau Sebé-Pedrós). Our thanks to the Node for letting us share it with you here.
This is the first paper from Jordi’s new lab at Oxford Brookes – more coming soon. And we will be soon recruiting a bioinformatician. Please contact Jordi Solana if this sounds like you!

References:
- Baguñà, J. and Romero, R., 1981. Quantitative analysis of cell types during growth, degrowth and regeneration in the planarians Dugesia mediterranea and Dugesia tigrina. Hydrobiologia, 84(1), pp.181-194. https://link.springer.com/article/10.1007/BF00026179
- David, C.N., 1973. A quantitative method for maceration of Hydra tissue. Wilhelm Roux Archiv für Entwicklungsmechanik der Organismen, 171(4), pp.259-268. https://link.springer.com/article/10.1007%2FBF00577724
- García-Castro, H., Kenny, N.J., Álvarez-Campos, P., Mason, V., Schönauer, A., Sleight, V.A., Neiro, J., Aboobaker, A., Permanyer, J., Iglesias, M. and Irimia, M., 2020. ACME dissociation: a versatile cell fixation-dissociation method for single-cell transcriptomics. bioRxiv. https://www.biorxiv.org/content/10.1101/2020.05.26.117234v
- Plass, M., Solana, J., Wolf, F.A., Ayoub, S., Misios, A., Glažar, P., Obermayer, B., Theis, F.J., Kocks, C. and Rajewsky, N., 2018. Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science, 360(6391). https://science.sciencemag.org/content/360/6391/eaaq1723
- Rosenberg, A.B., Roco, C.M., Muscat, R.A., Kuchina, A., Sample, P., Yao, Z., Graybuck, L.T., Peeler, D.J., Mukherjee, S., Chen, W. and Pun, S.H., 2018. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science, 360(6385), pp.176-182. https://science.sciencemag.org/content/360/6385/176
- Stuart, T. and Satija, R., 2019. Integrative single-cell analysis. Nature Reviews Genetics, 20(5), pp.257-272. https://doi.org/10.1038/s41576-019-0093-7


 (8 votes)
(8 votes)