A gnawing question: which cells are responsible for tooth renewal and regeneration?
Posted by Amnon Sharir, on 23 October 2019
By Amnon Sharir (UCSF), Allon M Klein (HMS), Ophir D Klein (UCSF)
As most mouse geneticists know, treating a rodent with malocclusion of its front teeth can be a Sisyphean task: as much as one trims the tooth, it grows right back within a few days (Figure 1). The basis of this often-frustrating situation is that rodent incisors contain a group of adult stem cells (SCs) at the base of the tooth that drive continuous growth of the organ. These SCs produce a constant supply of new cells that replace the cells that are lost from the tip of the tooth due to normal wear or tooth breakage. In contrast, human teeth are quite different: once they are mature, they no longer grow. As a result, wear and tear of the enamel, the hard layer that covers the teeth, as well as diseases like caries that disrupt the enamel, cannot be self-repaired.
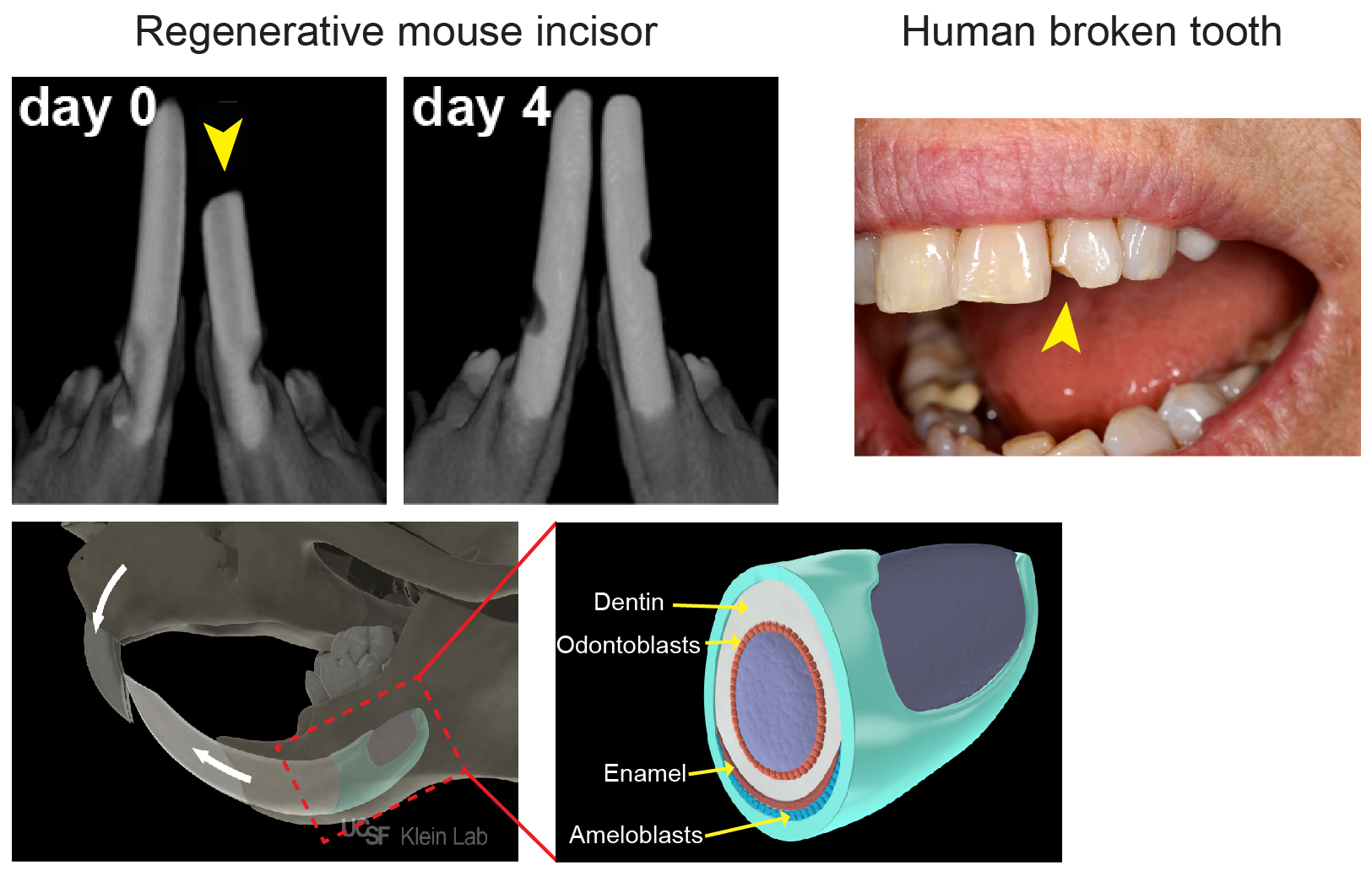
Because of their amazing regenerative abilities, our lab (klein.ucsf.edu), along with a number of others, has become very interested in mouse incisor SCs. We believe that understanding the mechanisms by which animals like mice normally renew their teeth will enable us to lay a foundation for human tooth regeneration. The mouse incisor provides a powerful platform for uncovering cellular behaviors, signaling pathways and transcriptional interactions that govern self-renewal and differentiation, and these findings have important implications for the field of SC biology. Most of the discoveries in the incisor epithelium, the tissue which produces the ameloblasts that lay down enamel (Figure 1), have resulted from candidate approaches. These studies identified several genes expressed by cells in the most proximal region of the incisor epithelium that give rise to the differentiated ameloblasts over a long period of time. Of note, these markers also largely, but not exclusively, co-localize with cells that are slowly cycling and therefore retain a label for long periods (label-retaining cells, LRCs). These experiments led to the widespread notion in the field that the SCs would be found among the LRCs. However, a number of key questions have remained unanswered, such as: Where exactly do the SCs in the tooth reside? How are SCs able to produce the correct number of ameloblasts to keep the tooth at a fixed length? How do SCs react to injuries such as trimming? Is there only one type of SC, or are there several discrete populations with variable functions?
Our adventure started back in 2013, when Allon Klein (klein.hms.harvard.edu) visited UCSF to share the quantitative approaches that his lab at Harvard was taking to study SC behavior. We realized immediately that these methods would be valuable for the mouse incisor field. The large, single-output incisor niche provides a contrast with the multiple, small, independent units found in other well-studied epithelial systems, such as the hair follicle and gut crypt. As such, it offers a rare opportunity to study the mechanisms that regulate cell number, as we can count the output of all of the SCs into the entire organ. The large size of the niche also offers the chance to study the unique challenges of maintaining proper SC numbers; for example, if a few crypts or hair follicles are lost, it is not a catastrophe for the animal, but loss of even one incisor SC niche would be fatal in the wild. Also, because teeth are largely dispensable for the survival of laboratory-housed mice, we can injure them and study their regeneration over extended periods without killing the animal.
At least initially, we assumed that our undertaking would involve a relatively straightforward analysis, as we thought that we knew the location of the SCs and their proliferation dynamics, and we had in our hands several inducible genetic tools for in vivo lineage tracing of these cells. As with many scientific adventures, however, things were more complicated than they seemed at first.
Historically, a classical view in the SC field in general has been that tissue SCs are slow cycling LRCs, and this was also true in the incisor field. While this view has evolved over the last decade, it is still often thought that SCs cycle less frequently than their immediate progeny, typically known as transit amplifying cells. In previous studies conducted by our lab and others, we pulsed mice with 5-bromo-2′-deoxyuridine (BrdU) to label proliferating cells during the early postnatal period and then aged the mice for several weeks. Cells within the presumed SC region at the base of the tooth, called the outer enamel epithelium, acquired BrdU label and retained it for several months. The extended retention of the BrdU label was interpreted to mean that these cells are SCs. However, to our surprise, when we pulsed 8-week-old mice with BrdU, the outer enamel epithelium was not labelled at all (Figure 2). No matter how much BrdU (or EdU, 5-ethynyl-2′-deoxyuridine) we used, the cells in the LRC region that we had presumed to contain an active SC population remained unlabeled! Lack of cell cycling in this region did not initially make sense to us, because the incisor fully turns over every 4-6 weeks, and therefore, we predicted the cycling time of the SCs to be relatively short — similar to that in other fast-cycling tissues, such as the epidermis and the intestinal epithelium.
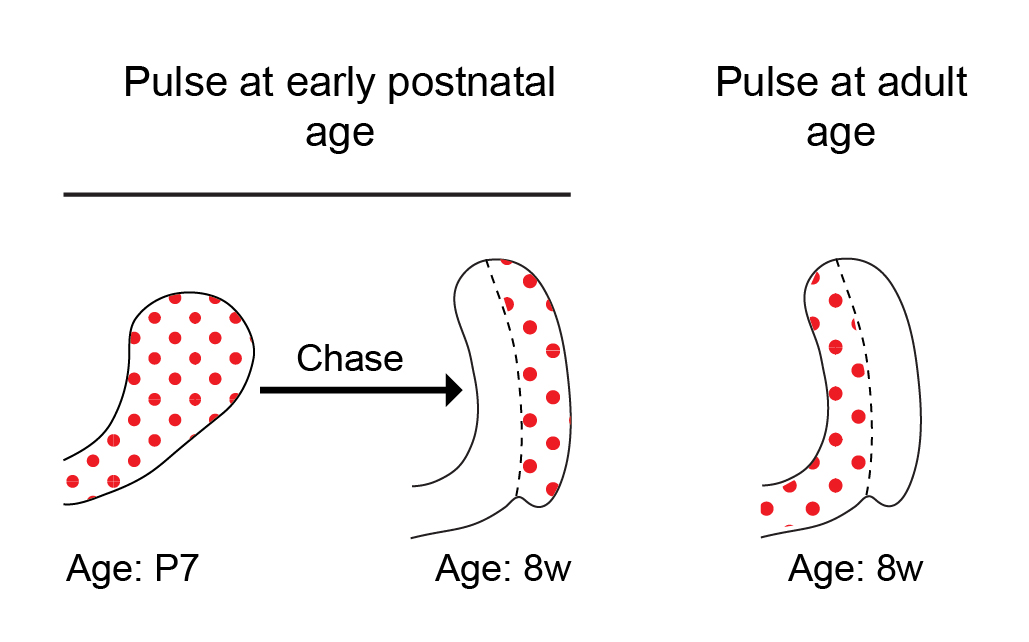
The dramatic transition from active proliferation to a dormant outer enamel epithelium over the first few weeks of the mouse’s life prompted us to define the time period during which the incisor is in steady state. To this end, we performed a series of 3D micro computed tomography (µCT) analyses of the incisor during postnatal development. We found that the incisor SC niche, called the labial cervical loop, is in steady-state between 8 and 16 weeks of age. During this period, the incisor growth rate is minimal, the cervical loop size is constant and proliferation is stable. We reasoned that the LRCs identified in previous incisor experiments represent post-mitotic cells that proliferate only during the postnatal expansion period, and we therefore focused our analysis on the steady state period.
Some cells must be cycling to incorporate a BrdU or EdU label. Since cells were not cycling in the outer enamel epithelium during our desired nucleotide pulse period, we decided to instead use an H2B-GFP label dilution system. In such a system, expression of inducible or repressible H2B-GFP, driven by a tet-response element, is controlled by a tetracycline (Tet)-transactivator (rtTA in Tet-On or tTA in Tet-Off). The proliferation dynamics of the organ can be tracked, because during the chase period, the resulting GFP is diluted by half during each cell division (Figure 3).
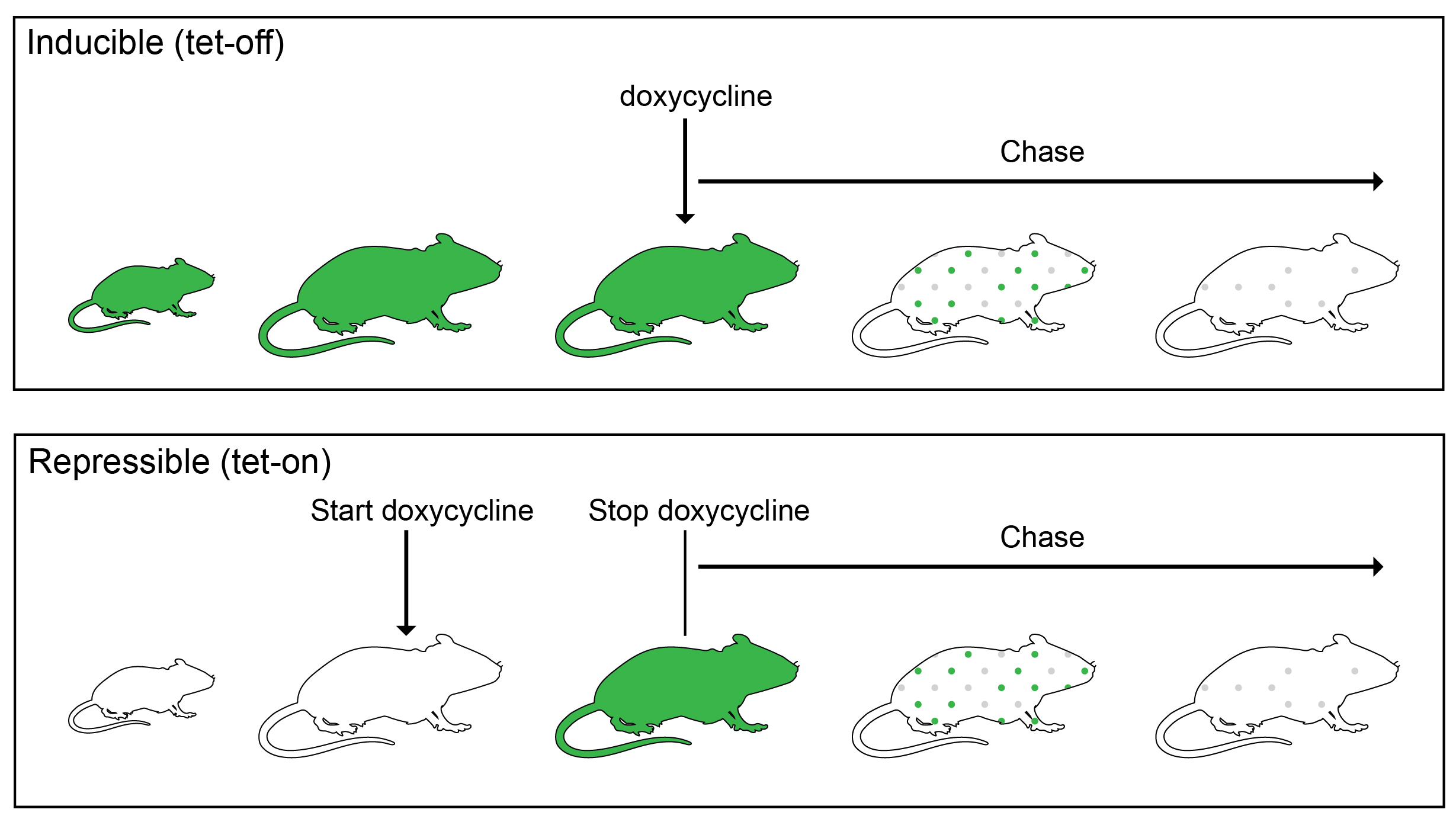
We initially used a K5tTa;tetOff-H2B-GFP, in which constitutive GFP protein expression in the incisor epithelium can be shut off by treatment with doxycycline1. However, we soon discovered that the incisors of the tetOff-H2B-GFP mice were abnormal: they were smaller, had cracks and frequently broke in our hands while we prepared them for analysis, and appeared chalky white (in mice, this is not a sign of good tooth hygiene, but rather an indication of lack of mineralization). Our μCT analysis confirmed that indeed there was a significant decrease in incisor volume and enamel density. So, this line could not be used to determine incisor cell kinetics (anomalies of other organs, such as the cornea, have been noted by others2, which perhaps is due to very high GFP levels in early life that are destructive to some organs). We then switched to the repressible TetOn-H2B-GFP line, in which H2B-GFP is activated by doxycycline treatment3. To our relief, the incisors of these mice appeared normal. However, again with this line, the outer enamel epithelium was devoid of labeling, unless we pulsed the mice during the perinatal period. A similar lack of cell labeling in other tissues, such as the olfactory bulb and the spinal cord, has been noted by the researcher who developed the line and was attributed to an inability of doxycycline to cross the blood-brain barrier4. In our case, we think that the cells are not labelled in the incisor because they are so quiescent that they don’t replace unlabeled histone H2B with the labeled one.
We decided to stick with the TetOn-H2B-GFP line, in which we were able to label the active region of the incisor epithelium and asses proliferation dynamics using a short pulse of doxycycline. We first used flow cytometry to sort the epithelial cells during a chase period and measure their GFP intensities, and then we modeled how the GFP signal distribution should change between chase day 1 to chase day 7 to infer two parameters: the fraction of cells that are proliferating, and their division rate. The model we specifically considered assumed that the number of divisions of any individual cell was Poisson-distributed, which gave a very good fit to the data. The best fit was when the average number of divisions in six days was around 3 (2.98 ± 0.20), and that initially 60±15% of cells were post-mitotic.
While the sorted GFP results provided us with valuable information regarding proliferation dynamics in the incisor epithelium, these data did not tell us anything about the spatial distribution of division. We decided to investigate the proliferation dynamics of the entire organ in situ. Using a two-photon microscope, we acquired images of the entire proximal region of the incisor at 45 minutes and 48 hours after EdU injections. We chose 48 hours, because at this time-point the cells have divided once on average, and no cells have yet been lost due to distal movement along the incisor length. It was clear from looking at the images that many EdU cells moved from the active proliferative region to other areas within the cervical loop. However, quantitative analysis of the location and level of EdU in each cell remained a challenge. This was especially true in our enormous region of interest, which contained about 175 images of 900 µm (length) X 900 µm (height) X 350 µm (width), with very densely packed cells and variabilities in laser intensity, due to the need to penetrate deeper sections of the sample. At this point, we were introduced by our colleague Jacqui Tabler to Kyle Harrington (kyleharrington.com), an expert in image analysis from the University of Idaho, who ultimately developed a segmentation pipeline tailored to our needs.
The software that Kyle developed uses a region competition algorithm to express image intensity and statistics of candidate regions as energy terms that are iteratively refined and balanced until the segmentation converges. After filtering regions based upon the expected size of cells, it measures the EdU signal at the center of the cell to determine proliferation status (Figure 4). The result of this image analysis pipeline was a 3D map of the spatial distribution of individual cells and their corresponding EdU signal, which confirmed our observations from the thin sections and fit well with our single cell RNA sequencing data regarding the sites of active cell division and the flow of cells from these sites to other regions in the incisor epithelium.
Figure 4: An example of the segmentation pipeline shown in video which was rendered in the freely-available ImageJ-based tool, SciView5. Cell membrane labeled in magenta and the center of segmented EdU+ cells shown in yellow.
In this blog post, we have shared several aspects of our thought process during our recently-completed project, in the hope that other researchers will find it useful when planning similar experiments or facing unexpected outcomes in their analyses of epithelial SCs. In our recent paper6, we integrate our quantitative proliferation kinetics with unbiased single cell RNA-seq analysis and genetic lineage tracing. We were able to uncover cell behaviors that upended the reigning dogma about the identity, location and function of progenitor cells in our tissue of interest.
References:
- Tumbar, T. et al. Defining the epithelial stem cell niche in skin. Science 303, 359–363 (2004).
- Sartaj, R. et al. Characterization of slow cycling corneal limbal epithelial cells identifies putative stem cell markers. Sci. Rep. 7, 3793 (2017).
- Foudi, A. et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 27, 84–90 (2009).
- Brennand, K. et al. All β Cells Contribute Equally to Islet Growth and Maintenance. PLOS Biol. 5, e163 (2007).
- Günther, U. et al. Scenery–Flexible Virtual Reality Visualization on the Java VM. Accepted IEEE VIS(arXiv:1906.06726 (2019)).
- Sharir, A. et al. A large pool of actively cycling progenitors orchestrates self-renewal and injury repair of an ectodermal appendage. Nat. Cell Biol. 21, 1102–1112 (2019).


 (5 votes)
(5 votes)