An interview with Guillermo Serrano Nájera, BSDB Beddington Medal winner 2022
Posted by Helen Zenner, on 14 June 2022
The Beddington Medal is awarded by the BSDB for the best PhD thesis in developmental biology, defended in the year prior to the award. The medal is named in memory of Rosa Beddington, who made major contributions to both the field of developmental biology and the BSDB. The artwork on the medal is from Rosa’s own drawings.

In 2022, the medal was awarded to Guillermo Serrano Nájera. Guillermo completed his PhD in Cornelis Weijer’s lab in Dundee and he presented his work at the recent BSCB/BSDB annual meeting in Warwick. I caught up with Guillermo over Teams to find out more about his career so far.

Where are you originally from?
I’m from Spain, from Madrid. I studied biochemistry at the university there. After completing my undergraduate degree, I decided I wanted to do something more interdisciplinary, so I stayed on to do a master’s degree in biophysics. At this point, I moved to Newcastle in the UK to do a second master’s degree in computational biology, but my main focus for this move was to improve my English. I could read very well but wanted to improve my speaking skills to be able to communicate more easily in science.
When did you first become interested in science?
I’ve been very interested in science for a long time, since childhood really. But I was also interested in just about everything! I liked archaeology, anthropology, literature, even philosophy. Then little by little, I became more convinced about focussing on science. Before university, I was choosing between physics and biology. I like the methods of physics, but ultimately decided to go along the biology route because I’m more interested in asking questions about living things. That’s why ultimately, I wanted to study science, and biology in particular.
How did you come to do a PhD in Cornelis (Kees) Weijer’s lab?
I was completely convinced that I wanted to study developmental biology for my PhD. I had wanted to study developmental biology for a long time, but for one reason or another, I couldn’t join a suitable lab. The second factor in choosing my thesis lab was that I wanted to do something interdisciplinary, using computing, modelling, as well as biology. Of course, there are many labs that are interdisciplinary, but I found that most of them were looking for biologists and physicists to collaborate in the lab, rather than truly interdisciplinary researchers. But I told Kees, my supervisor, that I wanted to work on all aspects of our research. He told me, ‘of course, there’s no other way!’ So, we were a match, and ended up working together.
Tell us about your PhD project: what were the main questions you were trying to answer?
The main question that I wanted to answer was, how are cell behaviours coordinated during animal development to generate shapes. We use chick embryo gastrulation as our model, and in this continuous epithelial layer, the cells can only have four different behaviours. This makes things easy, right! The cells can rearrange or interchange neighbours, they can change shape, they can divide, or they can ingress and abandon the layer. All these four cell behaviours coordinate in space and time over thousands and thousands of cells to generate the primitive streak during avian gastrulation. My question was, how do all these cells know what to do in each moment during this process. I used chemical perturbations to disrupt different cell behaviours as this was an easy method to combine with imaging. And the surprise was that by disrupting these behaviours, we generated structures that are very similar to gastrulation in other organisms. After these initial observations, we began to focus on our interpretation of these results in our next experiments. But the original question was, how are cell behaviours coordinated during development.
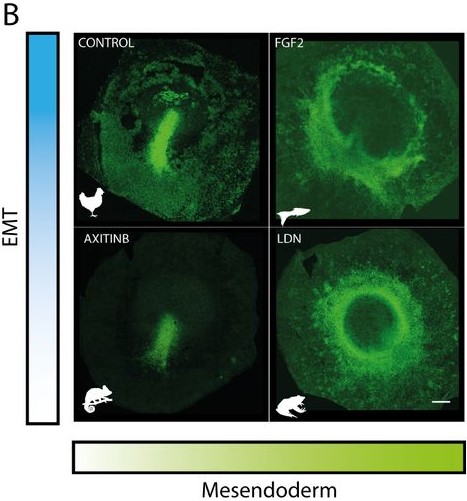
You have two bioRxiv preprints arising from this work, where you use both experimental data and modelling to address the differences in gastrulation in vertebrates, can you tell us about this collaboration and your results?
The biology paper is mostly my own work from my PhD, where we used chemical perturbations to change two parameters during gastrulation in the chick embryo; one was the amount of mesoderm, and the other was whether the cells in the mesoderm territory can undergo cell ingress. We found that by changing just these two parameters, you can generate different morphologies in the embryo, which look very similar to gastrulation in other organisms. At the same time, we observed myosin cables in the mesoderm territories, and we think these cables are producing the force to move the cells. For a long time, Kees had thought that these cables could be mechanosensitive, and that tension helps with their assembly. This could explain the movement of the cells and the self-organisation of the tissue flows. He was looking for collaborators to try to explore this in a more formal way using mathematical models. Kees was meeting with Mattia Serra from the Mahadevan lab about another project, and he asked if they would be interested in collaborating on this research. They began working on the model and how to implement it. I joined them because I had a way to test the model experimentally. We introduced the same perturbations into the model and were able to reproduce the experimental results. So, it was a confirmation that this principle of myosin self-organisation, and the resulting tissue flows could be underlying the structures that are formed during gastrulation.
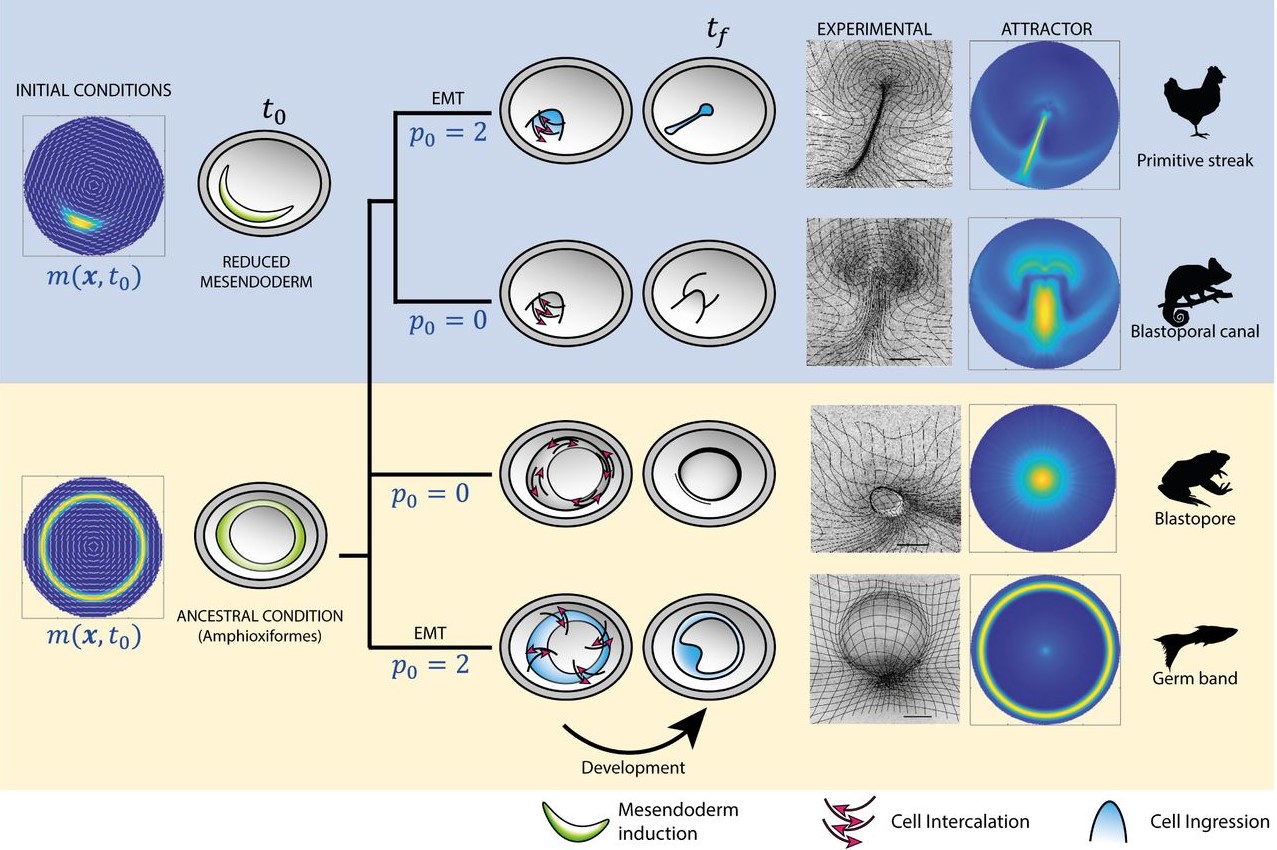
It must have been exciting when you saw that you could phenocopy the different types of gastrulation from other organisms.
In the beginning, we were not thinking about that at all. Actually, the first treatment that I performed resulted in an embryo that looked similar to the gastrulation in reptiles, with a small invagination in front of the primitive streak. We had that result for a long time before we thought about it from that perspective. Then, after reading papers about gastrulation in chameleons, I told Kees that I thought that our result looked similar to gastrulation in reptiles. We knew about the paper from Alev Cantas, Guojun Sheng and colleagues, where they create a circular primitive streak in chick embryos using FGF. They compare this to the circular primitive streak in reptiles, and the evo-devo perspective that they had in their paper was super inspiring for me. They called this circular primitive streak a blastopore, and in the beginning that is what we were calling it. But later, one of our treatments caused a complete invagination of the epiblast and we realised that our first result was not the blastopore! It was very, very exciting, especially when this last invagination happened and we realised that we could phenocopy all the gastrulation modes, and we began to interpret all our results in a new light.
If you took one abiding memory with you from your PhD, what would it be?
I had a lot of fun during my PhD, so choosing one moment is hard! I think I will have to choose an everyday moment that I always enjoyed. So, I have seen the movement of cells and the formation of the primitive streak thousands and thousands of times, but each time, after doing my experiments, I loved looking at the movies! I find the process of gastrulation very mysterious and beautiful, and that hasn’t changed since my first experiments. And I think that kept me going a lot – just to feel that mystery.
You been involved in a number of different projects including publishing a paper on ‘Trendygene’, a pipeline for detecting targets for drug discovery, how did you come to work on this project?
In general, I fall in love with everything, that’s my problem! At the beginning of my PhD, I was very interested in artificial intelligence. It’s a fascinating topic, machine learning; I find it amazing. In my PhD programme, the BBSRC EASTBIO Doctoral Training Partnership, you have to complete an internship. I decided that I wanted to do an internship where I could learn some applications of artificial intelligence. I had already started studying by myself with a few online courses. And for my internship, I went to a pharmaceutical company called Exscientia. They wanted to be able to detect emerging areas in the biomedical literature to focus their efforts in producing new drugs. I came up with this idea, which we called Trendygene. I can briefly explain the background: we trained a deep neural network, to predict the number of publications about each gene in the human genome in a particular year, for example in 2020, but only using the data from 1980 to 2013. As well as publications, we incorporated data from recent reviews, citations, clinical trials, everything. And it works very well! But we were more interested in looking at when it fails, because in some cases, it fails very badly. These were unusual, because, in general, most of the trajectories of publications are very, very predictable – that’s why the machine learning approach works, but it makes huge mistakes for a small subset of genes. The interpretation for these genes is that something happened in the last few years, which are not included in the time series, that radically change the publication dynamics in unexpected ways. I think that our work is original because instead of using artificial intelligence to predict something, we’re interested in when it fails. The cases when it fails tend to be due to a recent discovery. We selected a couple of genes to talk about in the paper, and to say why we think they are trendy. One of them is PIEZO1, the mechanosensor which was recognised in the Nobel Prize in Physiology or Medicine awarded in 2021; we could detect it in July, which was very nice.
Did the network ever fail in the opposite direction?
We did find that we could have anti-trendy genes. In general, the network fails more often in the other direction; the anti-trendy genes tend to have smaller errors. In the first versions of the manuscript, we included some analysis of why some things are anti-trendy, but they were more difficult to analyse. Some examples of anti-trendy genes were genes that have become obsolete in diagnostics due to a new alternative. So, we could detect these genes, but they were not as interesting so we decided just to focus on what is trendy!
You also had a poster at the BSCB/BSDB meeting on self-domestication in naked mole rats – how did you start working on this project?
So, just before the pandemic, I became obsessed with domestication. I think it is a very interesting problem. Domestication is probably one of the longest running biology experiments that we have ever done! Which means we have all these results out there just ready for us to look at. One of the first scientists to look was, of course, Darwin. He wrote a massive book on the topic called ‘The Variation of Animals and Plants Under Domestication’. I went to the library in Dundee to try to borrow the book, and they told me that it would be a couple of days before I could collect it. When I arrived, I found that it was the first edition of the book! It had been in the basement of the library for 97 years before I got it. It is a beautiful book with original binding, it is incredible! I spent the lockdown with that book in my home library because the main library was closed.
So, you were allowed to take the book out of the library?!
Yes, I was. I actually wrote to them because, although the book was in the catalogue, I wasn’t sure that they realised what they had. They are now hoping to showcase the book at the library in the future. Anyway, as I said I became obsessed with domestication. One of the things that was most interesting to me is something called domestication syndrome, which is a series of traits, both morphological and behavioural that are characteristic of domestic animals but are not very common in wild species. The most famous example is drooping ears; only elephants have drooping ears in the wild, and they use them to control their body temperature. But all the other animals that you can think of with drooping ears are domesticated animals, dogs, pigs, cows. So, why do domesticated animals have drooping ears and while wild animals do not? Another famous example attributed to domestication syndrome are the white patches on the skin, like you see on cows. This kind of coloration is not present in the wild or is at least very uncommon. There are many other characteristic traits, for example a small snout and a reduction in the number and size of teeth. Interestingly, all these tissues, the teeth, the snout, the melanocytes responsible for the pigmentation, are derived from neural crest cells. The neural crest can generate very different tissues during development including the peripheral nervous system and the adrenal glands. That is really important because domestication syndrome theory says that domesticated species have a reduced fear response, they don’t have a fight or flight response. This is why you can pet a cow and it will not attacked you or run away. So, if you have problems in your neural crest cells, and they cannot migrate well or proliferate well or differentiate well, then they you will have a reduced peripheral nervous system, which transmits the fear response from the brain to the body and you will have lower adrenaline levels because of the effect on adrenal gland and so on. And as a side effect, these defective in the behaviour of the neural crest cells, will generate problems in other tissues such as the cartilage of your ear, or with your teeth etc. It’s just a theory, but I think is really beautiful. It fits with many animals, although it’s probably not the only way to domesticate. And then another one of these domestication syndrome traits is to be fully naked. For example, there are naked dogs, naked cats, naked horses (which looks extremely bizarre), pigs, and of course the naked mole rat. I was really interested in the naked mole rat because they are eusocial animals, they they behave like bees, with a queen and then other workers. There are rat miners, and rat nurses, I’m oversimplifying of course! But basically, they have this kind of social organisation. So, it occurred to me that, and it has also proposed in the literature, that animals could self-domesticate to live in groups, to increase their social tolerance. This led me to think that maybe the naked mole rat is a self-domesticated animal that self-domesticated to improve its social tolerance and that’s how they managed to evolve this eusocial behaviour. They live in very large groups of up to 300 individuals, which is unusual.
I began to look at this project with a friend, Koryu Kin, during the pandemic. We met in the street as I was wheeling my lab chair down the street to my home and Koryu was on the way to the lab to collect his things at the beginning of the lockdown. We had discussed the naked mole rat project before and when me met, Koryu told me, maybe this is the moment to work on it. He was right! We are now trying to publish our paper, where we defend our hypothesis that the naked mole rat presents self-domestication syndrome traits, and that this could improve the social tolerance and be related to the evolution of eusociality in these animals. We also present a hypothesis that the eusocial behaviour is also linked with the properties of the soil.
Sticking with side-projects that you have done during your PhD, you were selected for the Roche Continents programme which brings together artists and scientists, can you tell us more about how you became involved in the programme?
During my PhD, I have been involved in mixing science with art. For example, I am very interested in computational art. In the past, I created a couple of bots online, for example Twitter bots, which detect haiku, Japanese poems with a very particular set of syllables. The bot I created detects when something written in Spanish can be cut in such a way to reproduce a haiku. Basically, the bot retweets you to say ‘hey, you wrote a haiku, though you probably didn’t know it!’ While in Dundee, I’ve also been a tutor for a programme where students in art are encouraged to link up with scientists, to find inspiration from science, and to work on a project together. Because of my interest in art and science, I decided to apply to Roche Continents, which is a programme that Roche organises where they bring together art students and science students for one week every summer in Salzburg. We did workshops together, and we also went to the opera and the theatre. It was very, very interesting and a great experience. I would recommend it to everybody!
What are you working on now, as a postdoc with Ben Steventon in Cambridge, and how does Cambridge compare with Dundee?
I only moved here two months ago so I have not really begun working on my project yet. But I was attracted to Ben’s lab because of the different model organisms they are working on. They work on chick, fish, and mouse gastruloids. I’m exploring all of these models with different people in the lab and collaborating on different projects. However, my intention is probably to move more towards the mouse gastruloids. And in terms of Cambridge, when I first moved here, I found it very busy, noisy and with cars everywhere! I was so use to Dundee, which is a quiet city with a fantastic river. I thought that it was a very good place to be during lockdown because nature is very close. I really enjoyed my life there. But now here in Cambridge, I really like the massive community of scientists associated with the university, there are physicists, engineers, biologists; you have world experts in every aspect of science. That is fantastic.
Longer term, do you know if you plan to stay in science?
I don’t really have long term plans. For the moment, I would like to continue in science, and in academia in particular. I think that the kind of questions I want to answer are a better fit for academic science. That’s at least my medium-term plan!
Where do you think developmental biology will be in ten years?
I would like to see us have a very good understanding of developmental principles so that we can begin to manipulate them. Now, for example, we have these in vitro systems like gastruloids and other types of organoids; we understand the some of the developmental principles, and we can guide the development, but in the future, we could begin to engineer development. We could manipulate these programmes to generate things that are not in nature, or to find new purposes for the in vitro models. I think this engineering will happen, but I don’t know if it will be within 10 years or later.
When you’re not in the lab, what do you do for fun?
As I mentioned, I like computational art, I do that a lot. I also like walking in the forest, cooking with my girlfriend and spending time with friends. I think I have a very normal life outside of the lab!


 (1 votes)
(1 votes)