Behind the paper story: TWISTED, the birth of a new gene
Posted by Nayelli Marsch Martinez, on 25 April 2023
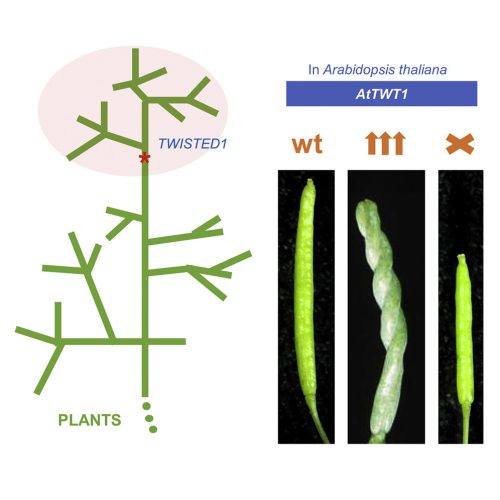
In the research paper by Nayelli Marsch-Martínez, Irepan Reyes-Olalde, Antonio Chalfun-Junior and colleagues, we report on the finding of a gene that appears to have been generated de novo, in a specific branch of the plant kingdom (Fig. 1). Some years ago, the idea of genes arising from non-genic sequences was thought to be highly unlikely, since genes were thought to arise from previously existing genes. This gene exquisitely modifies plant growth when overexpressed, causing twisted growth in many organs (Fig 1 or 2). Interestingly, even though this is a recent gene, its loss of function in the model plant Arabidopsis thaliana leads to developmental defects. There are very rare examples of new genes that influence development in such a clear way.
iScience: Twisting development, the birth of a potential new gene
How did you get started on this research?
This research started very long ago. It lasted almost 20 years, with pauses. Nayelli had moved from Mexico to The Netherlands to build an activation tagging population, as part of her Ph.D. (Marsch-Martinez et al., 2002). Many beautiful and interesting mutants were obtained, and other groups came to look for cool phenotypes. The twisted (twt) mutant was discovered by Jurriaan Mes when he came to screen the population and found a plant with siliques growing as helices. Since then, many people have worked on the mutant, first trying to identify the gene that was causing such a curious phenotype. When Nayelli, together with Stefan de Folter, moved to Mexico to work at CINVESTAV-IPN, the mutant came along, and the quest continued there.
What was already known about the twisted gene?
Actually, there was nothing known. We started by isolating the flanking region of the activation tagging element that was causing the phenotype. At that moment, the TWISTED gene was not even annotated. This caused a lot of trouble, because at the beginning, we tried to recapitulate the phenotype by overexpressing other genes, but never got the phenotype. Many constructs and transformants went by and we did not have the gene that was causing the twisted phenotype.
When doing the research, did you have any particular result or eureka moment that has stuck with you?
Yes, we were happy with the results of the recapitulation experiments, and it was very cool to finally see the recapitulation of the phenotype with the small gene. Also during this period we found that when a silencing construct was introduced into the activation tagging mutant, its phenotype was suppressed, which was exciting.
Another interesting moment was when we finally realized and accepted that this gene was not a common gene, and that probably was present only in a group of plants. This was hard to believe some years ago. A third moment was when the Cas9-edited plants showed phenotypes. We were not expecting that the loss of function of a taxonomically restricted gene would affect development like that. We thought it would maybe be more involved in coping with environmental conditions rather than development itself.
And what about the flipside: any moments of frustration or despair?
The first recapitulation experiments that Antonio (who did the initial experiments) tried, did not produce the phenotype, and this was very disappointing. He did a lot of work overexpressing different genes, analyzing their expression with marker lines and so on, but all the genes he worked on were not responsible for the phenotype. Another difficult moment was when we did the first BLAST searches and did not find any homologous sequences. At that time, these searches would produce zero hits. When Irepan, who also worked on the identification and characterization of the gene, presented the lack of hits in his master seminar, half of his classmates in the auditorium would tell him he was wrong. They could not believe that there was a gene that would not have any homologous sequences in the database. In recent years, we found hits to sequences only from species that are very close to Arabidopsis, but there was nothing at the time when we had just identified the gene.
It was also difficult to obtain a loss-of-function mutant. We ordered T-DNA insertional lines, but they did not cause knock-outs. One, where the T-DNA insertion was located in the UTR, even slightly increased the expression of the gene. Moreover, the phenotypes of silencing lines were not conclusive. It wasn’t until the CRISPR-Cas9 technology was available that we could have a real knock-out.
Where will this story take the lab?
For the moment, the different labs that participated in this study are working on other genes and subjects. We are also focusing on other genes, but we are still working on a side project in collaboration with Luis Delaye, one of our coauthors, to study the evolution of the gene in a more detailed manner. Nevertheless, it would be great to use this gene later in the future to modify fruit shape!
What next for you/your lab after this paper?
We continue to study fruit development and organ development in general, trying to discover new genes that participate in these processes.
Reference:
Marsch-Martinez, N., Greco, R., van Arkel, G., Herrera-Estrella, L., Pereira, A., (2002), Activation Tagging using the En-I maize transposon system in Arabidopsis, Plant Physiology, 129:1544-1556. https://doi.org/10.1104/pp.003327


 (No Ratings Yet)
(No Ratings Yet)